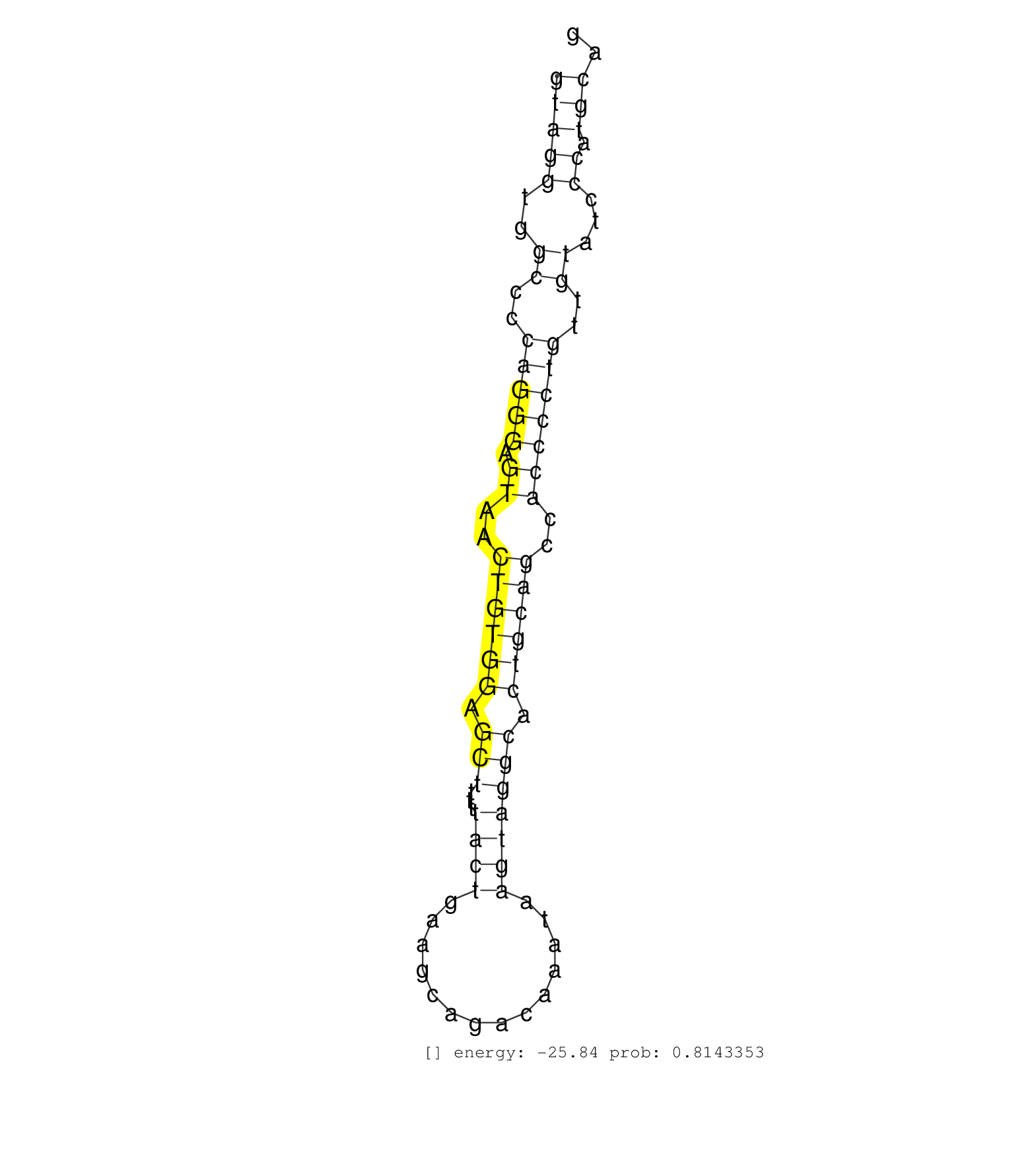

| Gene: Crtc2 | ID: uc008qbt.1_intron_5_0_chr3_90063415_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| TTATCCAGGCCCCACACCTCCCAGTGTCCTGCCCAGCCGCCGAGGAGGAGGTAGGTGGCCCCAGGGAGTAACTGTGGAGCTTTTTACTGAAGCAGACAAATAAGTAGGCACTGCAGCCACCCCTGTTGTATCCCATGCAGGTTTTCTGGATGGTGAGATGGATGCTAAAGGTATGTGTTCCTTACCAGCT ..................................................(((((..((..(((((.((..((((((.(((...((((..............))))))).))))))..)))))))..))...)).))).................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................GGGAGTAACTGTGGAGC.............................................................................................................. | 17 | 1 | 14.00 | 14.00 | 4.00 | - | 3.00 | 2.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TTCTGGATGGTGAGATGGATGCTAAAGtc.................. | 29 | tc | 3.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TTCTGGATGGTGAGATGGATGCTAAA..................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................TTCTGGATGGTGAGATGGATGCT........................ | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGGCCCCAGGGAGTAACTGTGG................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTTCTGGATGGTGAGATGGATGCTA....................... | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................AGTAACTGTGGAGCTTTTTACTGAAGC................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................TCTGGATGGTGAGATGGATGCT........................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TTTTTACTGAAGCAGACAAATAAGTAGGC................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTTTCTGGATGGTGAGATGGATGCTAAA..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................TCTGGATGGTGAGATGGggat......................... | 21 | ggat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................AGTAACTGTGGAGCTTTTTACTGAAGCAGAC............................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................GAGTAACTGTGGAGC.............................................................................................................. | 15 | 2 | 1.00 | 1.00 | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GGGAGTAACTGTGGAGCT............................................................................................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGGCCCCAGGGAGTAACTGggg................................................................................................................. | 27 | ggg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................CTGTGGAGCTTTTTACTGAAGCAGACA............................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................AGGGAGTAACTGTGGAGCT............................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGGATGGTGAGATGGATGCTAAAGtccc................ | 28 | tccc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTTCTGGATGGTGAGATGGATGCTAAAG.................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TTCTGGATGGTGAGATGGATGCTAAAGt................... | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................GTGGAGCTTTTTACTGAAGCAGACAAATAAG...................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................GTGAGATGGATGCTAAAG.................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| TTATCCAGGCCCCACACCTCCCAGTGTCCTGCCCAGCCGCCGAGGAGGAGGTAGGTGGCCCCAGGGAGTAACTGTGGAGCTTTTTACTGAAGCAGACAAATAAGTAGGCACTGCAGCCACCCCTGTTGTATCCCATGCAGGTTTTCTGGATGGTGAGATGGATGCTAAAGGTATGTGTTCCTTACCAGCT ..................................................(((((..((..(((((.((..((((((.(((...((((..............))))))).))))))..)))))))..))...)).))).................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|