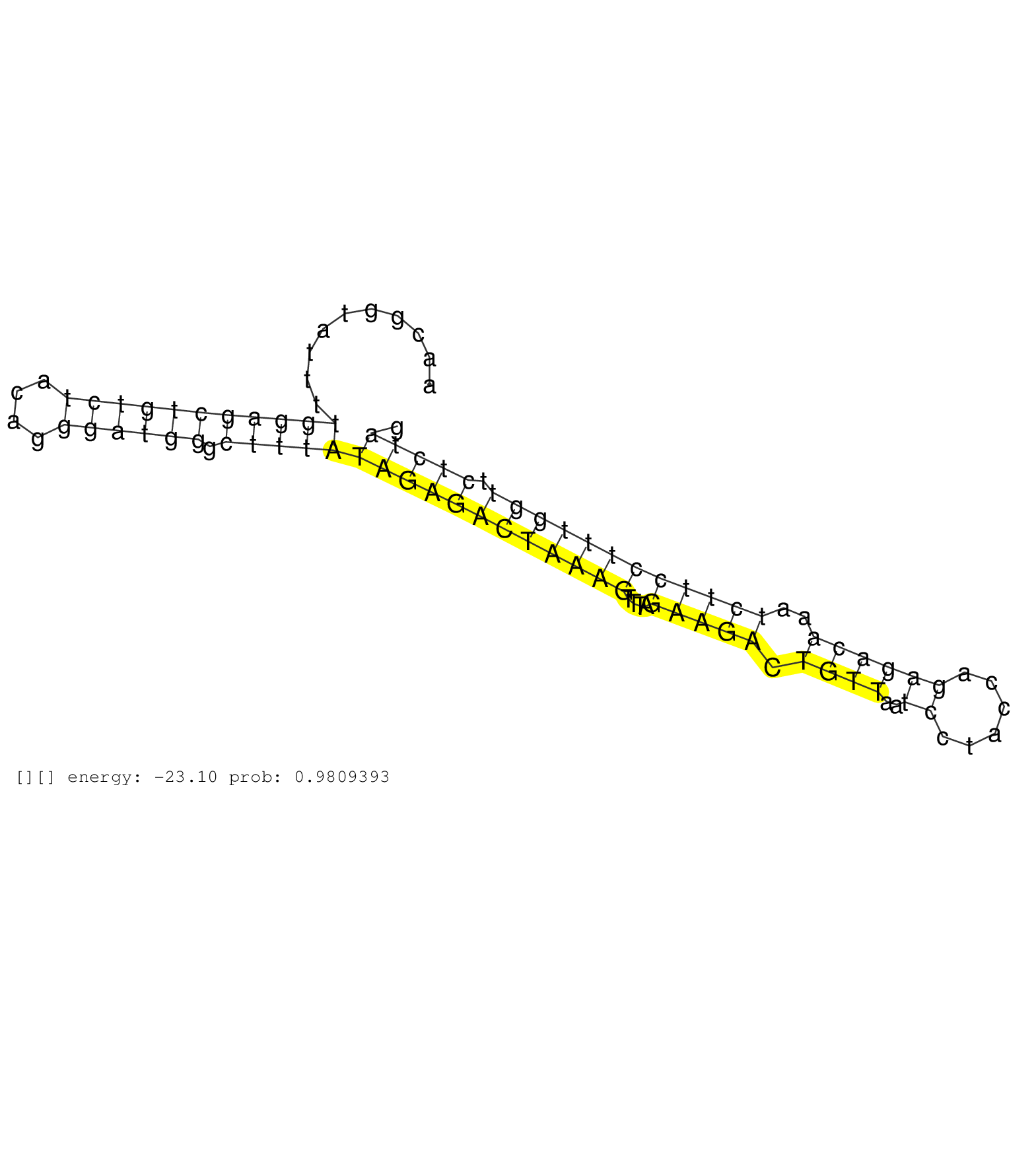

| Gene: AK019752 | ID: uc008qbk.1_intron_3_0_chr3_89922549_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(22) TESTES |
| GGGGGATTCAGAGTGAACTTCTGCTACGTTATTAGCAAGTTCAAGTTAGAGCCCAGCCTGGGTTACATAAGGCACTGTTTCAGAAAGCACACAAGCAAGCAACGGTATTTTGGAGCTGTCTACAGGGATGGGCTTTATAGAGACTAAAGTTTAGAAGACTGTTAATCCTACCAGAGACAAATCTTCCTTTGGTTCTCTAGGATTCTGAAGTACTCGGAAGCCGAGTATTCGCCCCCTGACTATATAATTG ..............................................................................................................(((((((((((....)))))).)))))((((((((((((....(((((.((((..((......))))))..)))))))))))).)))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................ATAGAGACTAAAGTTTAGAAGACTGTT....................................................................................... | 27 | 1 | 8.00 | 8.00 | 5.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGTT....................................................................................... | 28 | 1 | 5.00 | 5.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................ATAGAGACTAAAGTTTAGAAGACTGT........................................................................................ | 26 | 1 | 4.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGTTAAT.................................................................................... | 31 | 1 | 4.00 | 4.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TAGAGACTAAAGTTTAGAAGACTGTT....................................................................................... | 26 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................TGGGTTACATAAGGCACTGTTTCAGAAAGCACACAAGCAAGCAACGGTATTT............................................................................................................................................ | 52 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGT........................................................................................ | 27 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGTTAA..................................................................................... | 30 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGTTA...................................................................................... | 29 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................ATAGAGACTAAAGTTTAGAAGACTGTTA...................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................ATAGAGACTAAAGTTTAGAAGACTGTTAAT.................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TAGAGACTAAAGTTTAGAAGACTGTTA...................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TTTATAGAGACTAAAGTTTAGAAGACTGTT....................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGAGACTAAAGTTTAGAAGACTGTTA...................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TAAGGCACTGTTTCAGAAAGCACACAAGC.......................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................................................TAGGATTCTGAAGTACTCGGAAG.............................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGTTga..................................................................................... | 30 | ga | 1.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGTTAt..................................................................................... | 30 | t | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TTATAGAGACTAAAGTTTAGAAGACTGTT....................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................ATAGAGACTAAAGTTTAGAAGACTGTTAA..................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGGATTCTGAAGTACTCGGAAGCC............................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGGATTCTGAAGTACact.................................. | 19 | act | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................TTATAGAGACTAAAGTTTAGAAGACTGTTAAT.................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGTTAATC................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TAGAGACTAAAGTTTAGAAGACTGTTAAT.................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................AGTTAGAGCCCAGCCTGctc........................................................................................................................................................................................... | 20 | ctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................................TAGGATTCTGAAGTACTCGGAAGCCG........................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGGATTCTGAAGTACTCGGAAGCCGt.......................... | 27 | t | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGTTta..................................................................................... | 30 | ta | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................TATAGAGACTAAAGTTTAGAAGACTGTTAATt................................................................................... | 32 | t | 1.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TAGAGACTAAAGTTTAGAAGACTGTTAATCC.................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGGGGATTCAGAGTGAACTTCTGCTACGTTATTAGCAAGTTCAAGTTAGAGCCCAGCCTGGGTTACATAAGGCACTGTTTCAGAAAGCACACAAGCAAGCAACGGTATTTTGGAGCTGTCTACAGGGATGGGCTTTATAGAGACTAAAGTTTAGAAGACTGTTAATCCTACCAGAGACAAATCTTCCTTTGGTTCTCTAGGATTCTGAAGTACTCGGAAGCCGAGTATTCGCCCCCTGACTATATAATTG ..............................................................................................................(((((((((((....)))))).)))))((((((((((((....(((((.((((..((......))))))..)))))))))))).)))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................GAAAGCACACAAGCAAcc........................................................................................................................................................ | 18 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................CCAGCCTGGGTTACAacaa....................................................................................................................................................................................... | 19 | acaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |