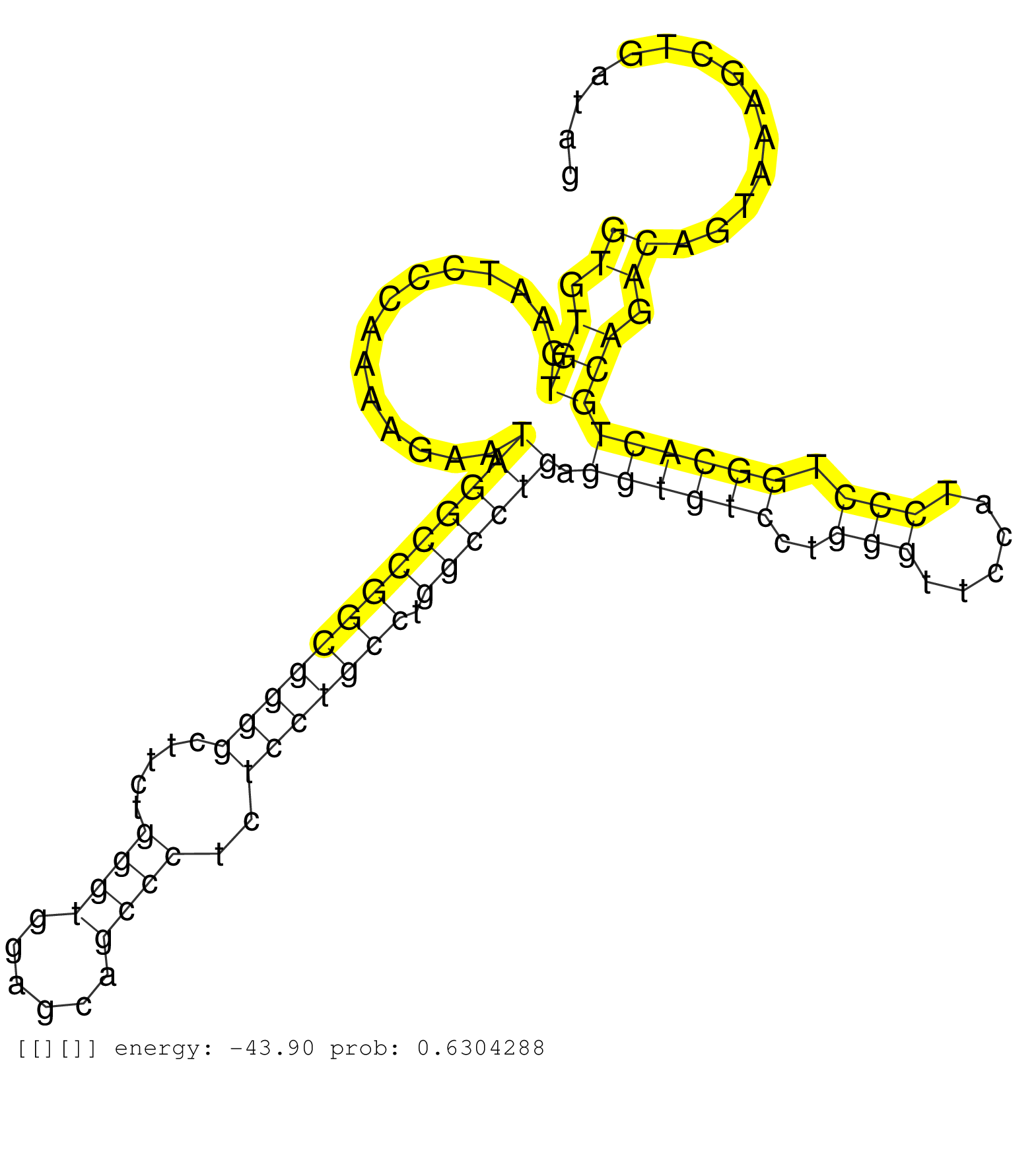

| Gene: Tpm3 | ID: uc008qbf.1_intron_2_0_chr3_89890502_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(15) TESTES |
| GCTTTGCAGAAGCTGGAGGAAGCAGAGAAGGCTGCTGATGAGAGTGAGAGGTGTGTGAATCCCAAAAGAATAGGCCGGCGGGGCTTCTGGGTGGAGCAGCCCTCTCCTGCCTGGCCTGAGGTGTCCTGGGTTCCATCCCTGGCACTGCAGACAGTAAAGCTGATAGTGGCTCTGACCTGGATTTTCCACTAGACCTGTTTTTTGTTTTGTTTGTGTTTTTTGTTTGAGGCATGGTCTCATGTAACCCAGA ..................................................((.(((..............(((((((((((((.....((((......))))..))))))).)))))).((((((..(((......))).))))))))).)).................................................................................................. ..................................................51.................................................................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGTGTGAATCCCAAAAGAATAGGCCGGC........................................................................................................................................................................... | 29 | 1 | 9.00 | 9.00 | - | 2.00 | 2.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .....GCAGAAGCTGGAGGAAGCAGAGAAGGCTGCTag.................................................................................................................................................................................................................... | 33 | ag | 7.00 | 0.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGTGTGAATCCCAAAAGAATAGGCCG............................................................................................................................................................................. | 27 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - |
| ..................................................GTGTGTGAATCCCAAAAGAATAGGCCGG............................................................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................TGTGTGAATCCCAAAAGAATAGGCCGGC........................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TTTTGTTTGTGTTTTTTGTTTGAGGC.................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TGCAGAAGCTGGAGGAAGCAGAGAA............................................................................................................................................................................................................................. | 25 | 5 | 1.20 | 1.20 | 1.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TCCCTGGCACTGCAGACAGTAAAGCTG........................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......CAGAAGCTGGAGGAAGCAGAGAAGt........................................................................................................................................................................................................................... | 25 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGTGTGAATCCCAAAAGAATAGGCCGGt........................................................................................................................................................................... | 29 | t | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TGGAGGAAGCAGAGAAGGCTGt....................................................................................................................................................................................................................... | 22 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................GTGTGAATCCCAAAAGAATAGGCCGGC........................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..TTTGCAGAAGCTGGAGGAAGCAGAGA.............................................................................................................................................................................................................................. | 26 | 5 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TCCCTGGCACTGCAGACAGTAAAGCTGt....................................................................................... | 28 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................TAAAGCTGATAGTGGCTCTGACCTGGA..................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGTGTGAATCCCAAAAGAATAGGCCGac........................................................................................................................................................................... | 29 | ac | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TTGCAGAAGCTGGAGGAAGCAGAGA.............................................................................................................................................................................................................................. | 25 | 5 | 0.80 | 0.80 | 0.80 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GAGAAGGCTGCTGATGAGAGTGAGA......................................................................................................................................................................................................... | 25 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - |
| ....TGCAGAAGCTGGAGGAAGCAGAGA.............................................................................................................................................................................................................................. | 24 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |
| .CTTTGCAGAAGCTGGAGGAAGCAGA................................................................................................................................................................................................................................ | 25 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - |
| ...TTGCAGAAGCTGGAGGAAGCAGAGAAGGCTGC....................................................................................................................................................................................................................... | 32 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - |
| .........AAGCTGGAGGAAGCAGAGAAG............................................................................................................................................................................................................................ | 21 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - |
| .....GCAGAAGCTGGAGGAAGCAGAGAAGG........................................................................................................................................................................................................................... | 26 | 5 | 0.20 | 0.20 | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - |
| .....GCAGAAGCTGGAGGAAGCAGAGAAGGCTGta...................................................................................................................................................................................................................... | 31 | ta | 0.20 | 0.20 | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - |
| .......AGAAGCTGGAGGAAGCAGAGAAGGCTG........................................................................................................................................................................................................................ | 27 | 5 | 0.20 | 0.20 | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - |
| .....................GCAGAGAAGGCTGCTGATGAGAGTG............................................................................................................................................................................................................ | 25 | 5 | 0.20 | 0.20 | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - |
| ....TGCAGAAGCTGGAGGAAGCAGAG............................................................................................................................................................................................................................... | 23 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - |
| .....GCAGAAGCTGGAGGAAGCAGAGAAGGCTG........................................................................................................................................................................................................................ | 29 | 5 | 0.20 | 0.20 | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TGCAGAAGCTGGAGGAAGCAGAGAAG............................................................................................................................................................................................................................ | 26 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - |
| ........................GAGAAGGCTGCTGATGAG................................................................................................................................................................................................................ | 18 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| GCTTTGCAGAAGCTGGAGGAAGCAGAGAAGGCTGCTGATGAGAGTGAGAGGTGTGTGAATCCCAAAAGAATAGGCCGGCGGGGCTTCTGGGTGGAGCAGCCCTCTCCTGCCTGGCCTGAGGTGTCCTGGGTTCCATCCCTGGCACTGCAGACAGTAAAGCTGATAGTGGCTCTGACCTGGATTTTCCACTAGACCTGTTTTTTGTTTTGTTTGTGTTTTTTGTTTGAGGCATGGTCTCATGTAACCCAGA ..................................................((.(((..............(((((((((((((.....((((......))))..))))))).)))))).((((((..(((......))).))))))))).)).................................................................................................. ..................................................51.................................................................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|