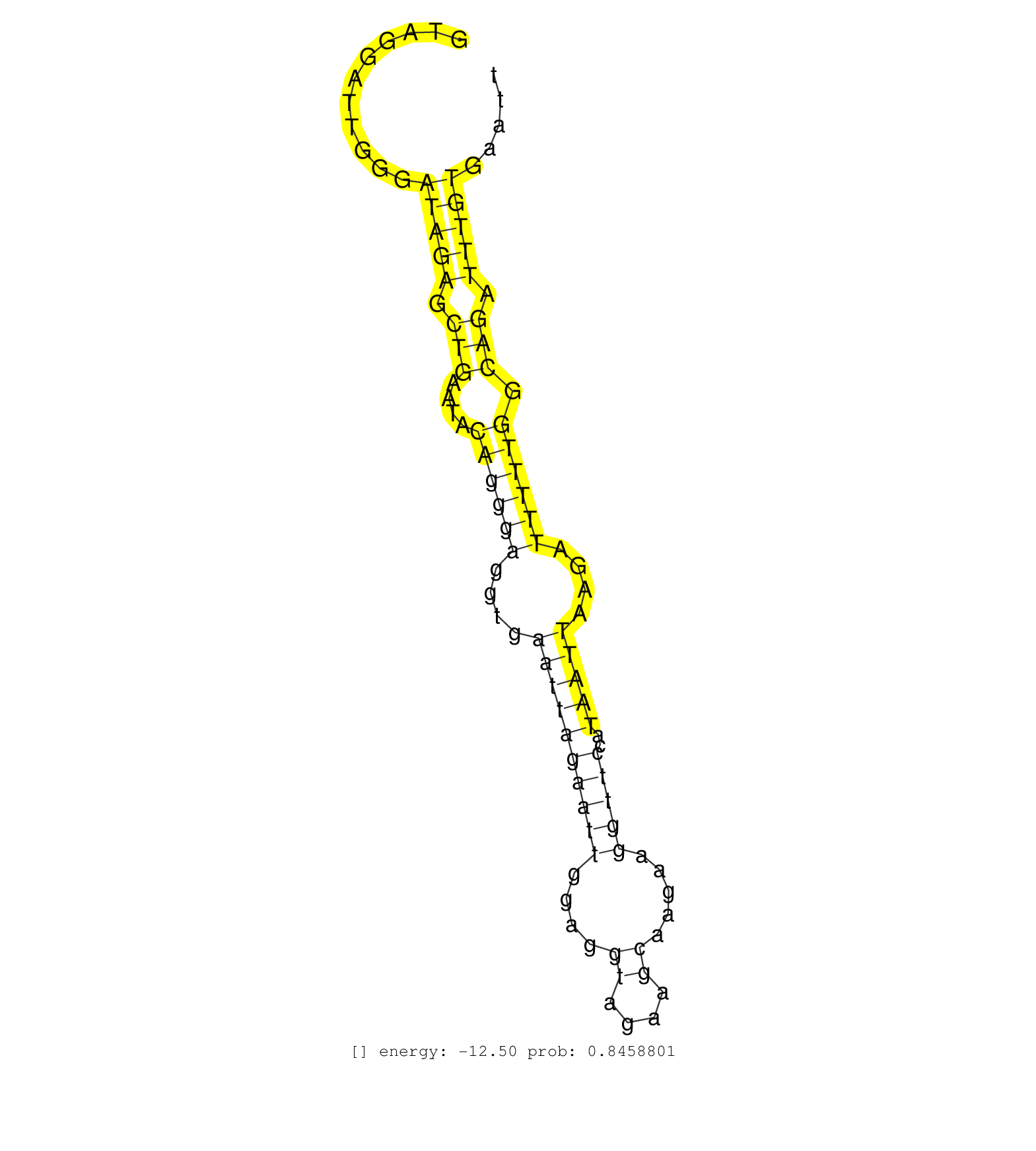
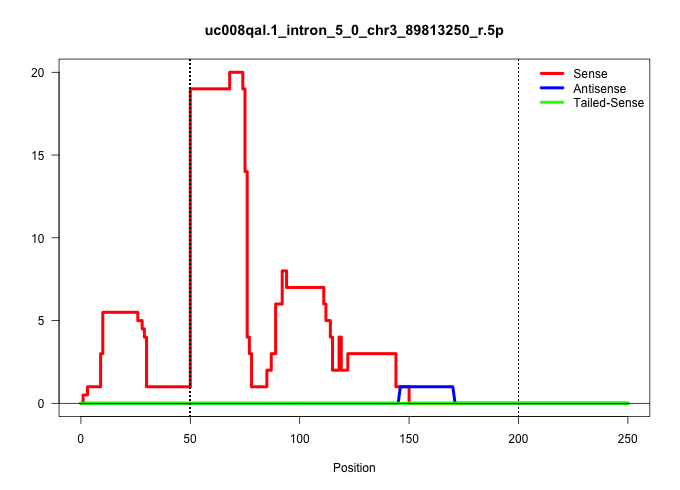
| Gene: Ubap2l | ID: uc008qal.1_intron_5_0_chr3_89813250_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| CACTCCACTTACTGGGAGGGATGGTAGCCTGGCCAGCAACCCTTACTCTGGTAGGATTGGGATAGAGCTGAATACAGGGAGGTGAATTAGAATTGGAGGTAGAAGCAAGAAGGTTCCATAATTAAGATTTTTGGCAGATTTGTGAATTTTCTTCCTCCCTACCCCTTGTTGTCATGGATATGATGAGTCAAGGCTGGGAGTTTCATAATTCTATTATGAATATGTTCCTGTGACATCATTTTTCTTTTAA .............................................................(((((.(((....((((((....((((((((((....((....)).....)))))..)))))....)))))).))).)))))........................................................................................................... ..................................................51...............................................................................................148.................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGATTGGGATAGAGCTGAATACA.............................................................................................................................................................................. | 26 | 1 | 10.00 | 10.00 | 5.00 | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTAGGATTGGGATAGAGCTGAATAC............................................................................................................................................................................... | 25 | 1 | 5.00 | 5.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TAATTAAGATTTTTGGCAGATTTGTG.......................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGATTGGGATAGAGCTGAATACAGG............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TTGGAGGTAGAAGCAAGAAGGTTCCAT................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........TACTGGGAGGGATGGTAGCCT............................................................................................................................................................................................................................ | 21 | 2 | 1.50 | 1.50 | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........ACTGGGAGGGATGGTAGCCT............................................................................................................................................................................................................................ | 20 | 2 | 1.50 | 1.50 | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGAATACAGGGAGGTGAATTAGAATT............................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................CTGAATACAGGGAGGacaa.................................................................................................................................................................... | 19 | acaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........ACTGGGAGGGATGGTAGCCTGGC......................................................................................................................................................................................................................... | 23 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 |
| .........................................................................................GAATTGGAGGTAGAAGCAAGAAGGT........................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................ATTGGGATAGAGCTGAATA................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................TAGAATTGGAGGTAGAAGCAAGAAGGTT....................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGATTGGGATAGAGCTGAATA................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CAGCAACCCTTACTCTG........................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................GAATTGGAGGTAGAAGCAAGAAG.......................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................GAATTGGAGGTAGAAGCAAGAAGGTT....................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................ATTAGAATTGGAGGTAGAAGCAAGAA........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAGGATTGGGATAGAGCTGAATACAG............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAAGATTTTTGGCAGATTTGTGAATTTT.................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .ACTCCACTTACTGGGAGGGATGGTAGC.............................................................................................................................................................................................................................. | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| .........TACTGGGAGGGATGGTA................................................................................................................................................................................................................................ | 17 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TCCACTTACTGGGAGGGATGGTAGCC............................................................................................................................................................................................................................. | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| CACTCCACTTACTGGGAGGGATGGTAGCCTGGCCAGCAACCCTTACTCTGGTAGGATTGGGATAGAGCTGAATACAGGGAGGTGAATTAGAATTGGAGGTAGAAGCAAGAAGGTTCCATAATTAAGATTTTTGGCAGATTTGTGAATTTTCTTCCTCCCTACCCCTTGTTGTCATGGATATGATGAGTCAAGGCTGGGAGTTTCATAATTCTATTATGAATATGTTCCTGTGACATCATTTTTCTTTTAA .............................................................(((((.(((....((((((....((((((((((....((....)).....)))))..)))))....)))))).))).)))))........................................................................................................... ..................................................51...............................................................................................148.................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TTTTCTTCCTCCCTACCCCTTGTTG............................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |