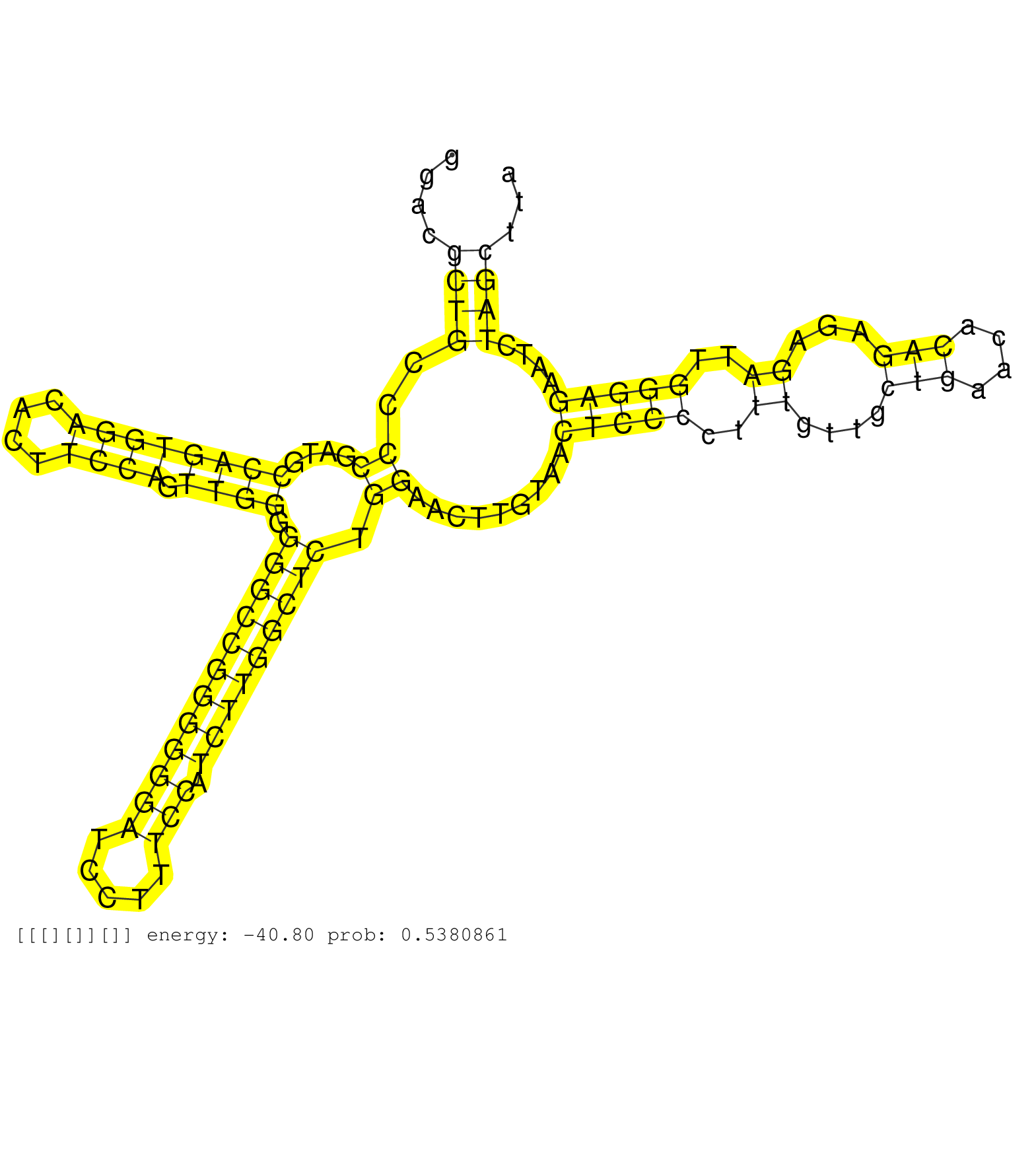
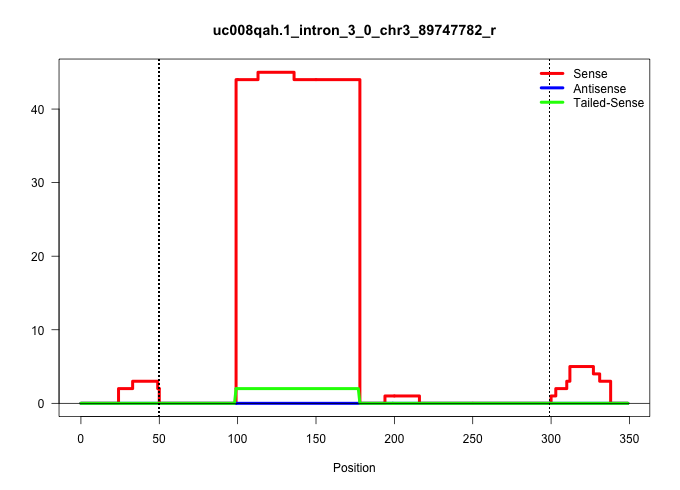
| Gene: Atp8b2 | ID: uc008qah.1_intron_3_0_chr3_89747782_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(13) TESTES |
| ACATCGTGTACACCTCCCTCCCCGTCCTAGCCATGGGCGTCTTTGACCAGGTATGGGGGTGGTCAGGGAAAGGTTGTCTGATATGCCAGGTGCGGGACGCTGCCCCGATGCCAGTGGACACTTCCAGTTGGGGGGCCGGGGGGATCCTTTCCATCTTGGCTCTGGAACTTGTAACTCCCCTTTGTTGCTGAACACAGAGAGATTGGGAGAATCTAGCTTAAGCCTGCATCTATTACCAAGACTTTAGGAAGGGGGACTGGAGAGCATCCGTGGCAGCTTCTGGGCTCCTCCCCTCACAGGATGTCCCGGAACAGCGAAGCATGGAGTACCCTAAACTGTACGAGCCAGG ..................................................................................................((((..((....((((((((....)))).)))).((((((((((((.....))).))))))))).)).........(((((..((....(((....)))...))..)))))....)))).................................................................................................................................... ..............................................................................................95...........................................................................................................................220............................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................CTGCCCCGATGCCAGTGGACACTTCCAGTTGGGGGGCCGGGGGGATCCTTTC...................................................................................................................................................................................................... | 52 | 1 | 49.00 | 49.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................AGGATGTCCCGGAACAGCG................................. | 19 | 1 | 8.00 | 8.00 | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................TAAACTGTACGAGCCAGG | 18 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ........................TCCTAGCCATGGGCGTCTTTGACCAG........................................................................................................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................AGCGAAGCATGGAGTACCCTAAACTG........... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................ATGTCCCGGAACAGCGAAGCATGGAGT...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................GTGGACACTTCCAGTTGGGGGGC..................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................................................................................................................................TCCCGGAACAGCGAAGCATGGAGTACCC.................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................TGGGCGTCTTTGACCA............................................................................................................................................................................................................................................................................................................ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................CAGAGAGATTGGGAGAATCTAG..................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................................................................................................................................................................................................................ACAGCGAAGCATGGAGTACCCTAAACTG........... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ACATCGTGTACACCTCCCTCCCCGTCCTAGCCATGGGCGTCTTTGACCAGGTATGGGGGTGGTCAGGGAAAGGTTGTCTGATATGCCAGGTGCGGGACGCTGCCCCGATGCCAGTGGACACTTCCAGTTGGGGGGCCGGGGGGATCCTTTCCATCTTGGCTCTGGAACTTGTAACTCCCCTTTGTTGCTGAACACAGAGAGATTGGGAGAATCTAGCTTAAGCCTGCATCTATTACCAAGACTTTAGGAAGGGGGACTGGAGAGCATCCGTGGCAGCTTCTGGGCTCCTCCCCTCACAGGATGTCCCGGAACAGCGAAGCATGGAGTACCCTAAACTGTACGAGCCAGG ..................................................................................................((((..((....((((((((....)))).)))).((((((((((((.....))).))))))))).)).........(((((..((....(((....)))...))..)))))....)))).................................................................................................................................... ..............................................................................................95...........................................................................................................................220............................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TCCATCTTGGCTCTGGAtgc....................................................................................................................................................................................... | 20 | tgc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................CAGTGGACACTTCCAtggg............................................................................................................................................................................................................................... | 19 | tggg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |