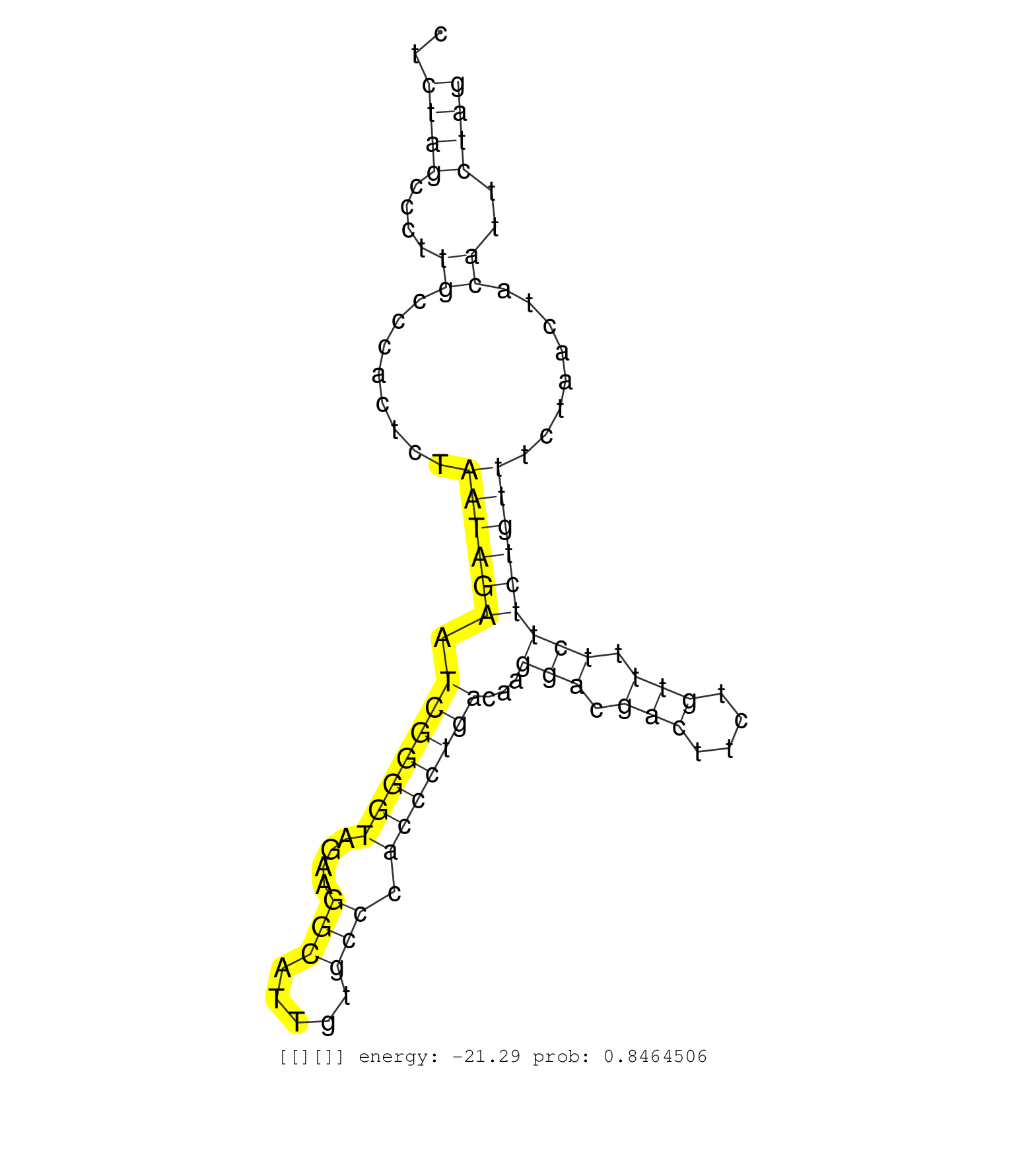
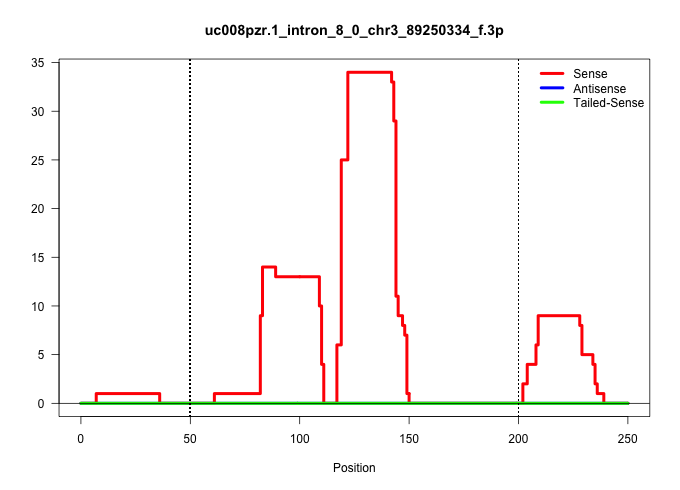
| Gene: Pbxip1 | ID: uc008pzr.1_intron_8_0_chr3_89250334_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| TGAAGAAGAGGAGGAACCCTTCTGGCTCTCCAGGCAGCACTGGGGCTCACACCCAGGGCCTTACACGCACTACACAGGCTCATTGCCAACAGAACTGCATCTCTAGCCCTTGCCCACTCTAATAGAATCGGGGTAGAAGGCATTGTGCCCACCCTGACAAGGACGACTTCTGTTTTCTTCTGTTTCTAACTACATTCTAGGCTCAGAAAGAAGAACTTCAGAGCCTGTTGCACCAGCCCAAAGGGCTGGA ......................................................................................................((((....((........((((((.(((((((....(((.....))).)))))))...(((.(((....))).)))))))))........))..)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................TAATAGAATCGGGGTAGAAGGCATT.......................................................................................................... | 25 | 1 | 16.00 | 16.00 | 6.00 | 1.00 | - | - | 3.00 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| ..........................................................................................................................TAGAATCGGGGTAGAAGGCATTGTGCC..................................................................................................... | 27 | 1 | 6.00 | 6.00 | - | 1.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TCTAATAGAATCGGGGTAGAAGGCAT........................................................................................................... | 26 | 1 | 4.00 | 4.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TTGCCAACAGAACTGCATCTCTAGCCCT............................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TGCCAACAGAACTGCATCTCTAGCCCT............................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TTGCCAACAGAACTGCATCTCTAGCCC............................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................................................................................GAAGAACTTCAGAGCCTGTTGCACCA............... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TCTAATAGAATCGGGGTAGAAGGCATT.......................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TTGCCAACAGAACTGCATCTCTAGCCCTT........................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TGCCAACAGAACTGCATCTCTAGCCCTT........................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................AGAAAGAAGAACTTCAGAGCCTGTT..................... | 25 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TAATAGAATCGGGGTAGAAGGCATTG......................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AGAAGAACTTCAGAGCCTGTTGCACCAG.............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGAATCGGGGTAGAAGGCATTGTGCCC.................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GAGGAGGAACCCTTCTGGCTCTCCAGGCA...................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TCAGAAAGAAGAACTTCAGAGCCTGT...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................................GAAGAACTTCAGAGCCTGTTGCACCAGCCC........... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TCAGAAAGAAGAACTTCAGAGCCTGTT..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................TTGCCAACAGAACTGCcttt.................................................................................................................................................... | 20 | cttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................AATCGGGGTAGAAGGCATTGTG....................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AGAAGAACTTCAGAGCCTGTTGCACC................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................TAATAGAATCGGGGTAGAAGGCA............................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................CACTCTAATAGAATCGGGGTAGAAGGCA............................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGAATCGGGGTAGAAGGCATTGTG....................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGAATCGGGGTAGAAGGCATTGTGC...................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TACACGCACTACACAGGCTCATTGCCAA................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGAAGAAGAGGAGGAACCCTTCTGGCTCTCCAGGCAGCACTGGGGCTCACACCCAGGGCCTTACACGCACTACACAGGCTCATTGCCAACAGAACTGCATCTCTAGCCCTTGCCCACTCTAATAGAATCGGGGTAGAAGGCATTGTGCCCACCCTGACAAGGACGACTTCTGTTTTCTTCTGTTTCTAACTACATTCTAGGCTCAGAAAGAAGAACTTCAGAGCCTGTTGCACCAGCCCAAAGGGCTGGA ......................................................................................................((((....((........((((((.(((((((....(((.....))).)))))))...(((.(((....))).)))))))))........))..)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................CTCTCCAGGCAGCACTgatt................................................................................................................................................................................................................. | 20 | gatt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CACCCTGACAAGGACtttt...................................................................................... | 19 | tttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |