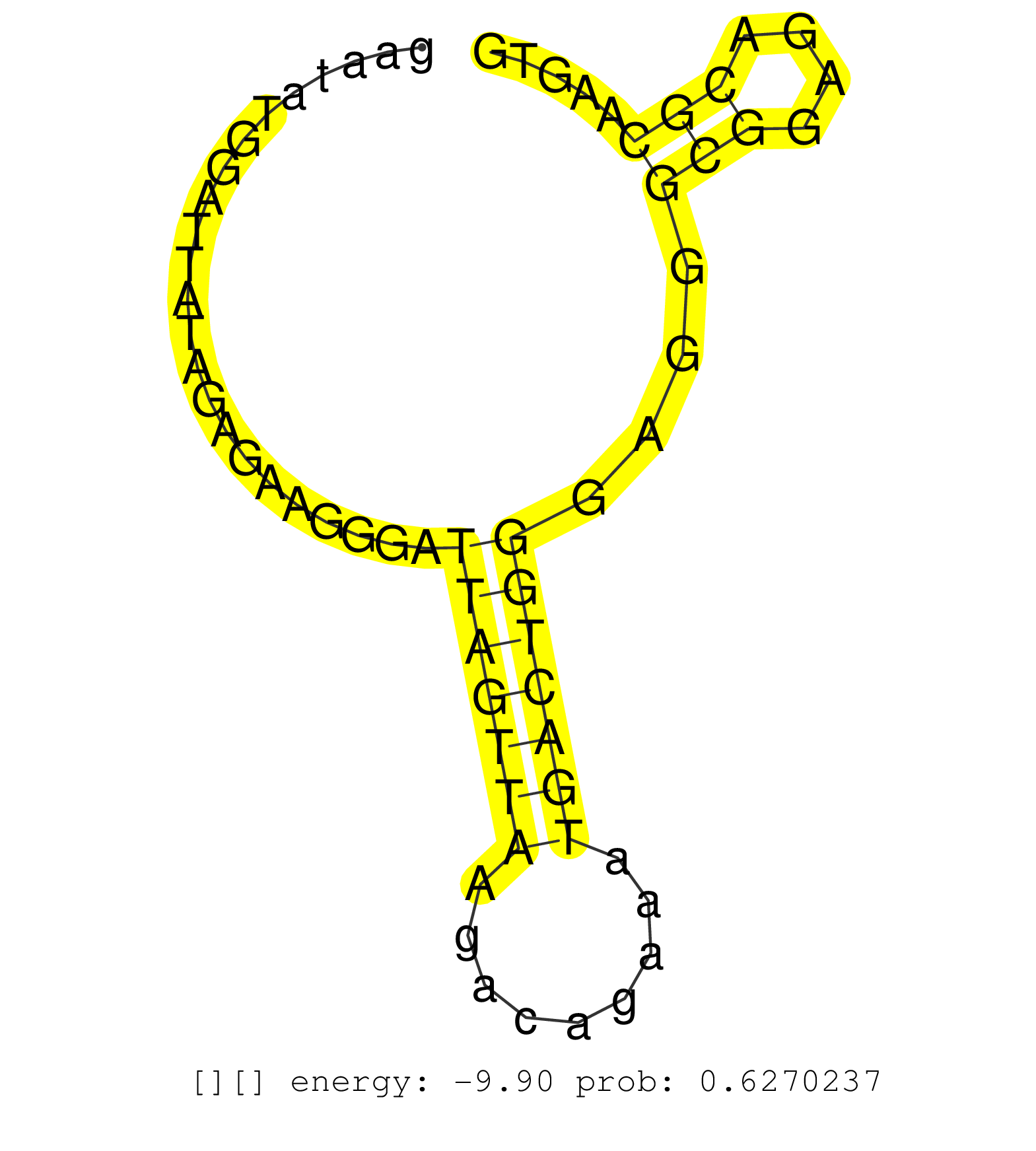

| Gene: Trim46 | ID: uc008pyk.1_intron_8_0_chr3_89048334_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| AGGATATGCAGACCTTCACTTCCATCATGGATGCACTAGTCCGCATCAGTGTGAGTTAAGGTGGGGTCAAGGGGAAGGGAGGGGATGTAAAGGGTCGGGGCAAGCCAGACCCTTGAACAGGTGGGGGTCCAGGAAGAATATGGATTATAGAGAAGGGATTAGTTAAGACAGAAATGACTGGGAGGGCGGAGACGCAAGTGTGGAGGGATCCTGGGGACAGCCGAGTGGTGAGACTCCCGTTTGCTGGGGT ..............................................................................................................................................................(((((((.........)))))))....(((....)))....................................................... .......................................................................................................................................136.............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR037901(GSM510437) testes_rep2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................AGTGTGGAGGGATCCaaa.................................... | 18 | aaa | 9.00 | 0.00 | 5.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGGATTATAGAGAAGGGATTAGTTAA.................................................................................... | 26 | 1 | 6.00 | 6.00 | - | 2.00 | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGACTGGGAGGGCGGAGACGCAAGTGT................................................. | 27 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................TAAGACAGAAATGACTGGGAGGGC............................................................... | 24 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................AGTGTGGAGGGATCtaaa.................................... | 18 | taaa | 3.00 | 0.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................AGAAATGACTGGGAGGGCGGAGACGC....................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TAAGACAGAAATGACTGGGAGGGCGG............................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................TATGGATTATAGAGAAGGGATTAGTT...................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGAGAAGGGATTAGTTAAGACAGA.............................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................AGTGTGGAGGGATCgaaa.................................... | 18 | gaaa | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTTAAGGTGGGGTCAAGGGGAA.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGACTGGGAGGGCGGAGACGCAAGTGa................................................. | 27 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................AGTGTGGAGGGATCagaa.................................... | 18 | agaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................GGAAGAATATGGATTATAGAGAAGGG............................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................TGGATTATAGAGAAGGGATTAGTTA..................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................GAAATGACTGGGAGGGCGGAGACGC....................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................................AGTGTGGAGGGATCCgaa.................................... | 18 | gaa | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTTAAGGTGGGGTCAAGGGGA............................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................AGTGTGGAGGGATCtaca.................................... | 18 | taca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................AGTGTGGAGGGATCCTat.................................... | 18 | at | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................GAAATGACTGGGAGGGCGGAGACGCAA..................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TATGGATTATAGAGAAGGGATTAGT....................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................TAAGACAGAAATGACTGGGAGGGCG.............................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ATGGATTATAGAGAAGGGATTA......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ATGGATTATAGAGAAGGGATTAGTTAA.................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................GGAAGAATATGGATTATAGAGAAGGGA............................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................AAGGGATTAGTTAAGACAGAAATGACTGGGAGG................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................TAGTTAAGACAGAAATGACTGGGAGGGC............................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TAGTCCGCATCAGTGTGAGTTAAGGTG........................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................AGTGTGGAGGGATCaaaa.................................... | 18 | aaaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................GCATCAGTGTGAGTTAAGG............................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................TAAGACAGAAATGACTGGGAGGGCGGt............................................................ | 27 | t | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGGATTATAGAGAAGGGATTAGTTAAG................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| AGGATATGCAGACCTTCACTTCCATCATGGATGCACTAGTCCGCATCAGTGTGAGTTAAGGTGGGGTCAAGGGGAAGGGAGGGGATGTAAAGGGTCGGGGCAAGCCAGACCCTTGAACAGGTGGGGGTCCAGGAAGAATATGGATTATAGAGAAGGGATTAGTTAAGACAGAAATGACTGGGAGGGCGGAGACGCAAGTGTGGAGGGATCCTGGGGACAGCCGAGTGGTGAGACTCCCGTTTGCTGGGGT ..............................................................................................................................................................(((((((.........)))))))....(((....)))....................................................... .......................................................................................................................................136.............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR037901(GSM510437) testes_rep2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................GGAGACGCAAGTGTGGAGGGATCCTGGG................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................CATCAGTGTGAGTTAAgat............................................................................................................................................................................................... | 19 | gat | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CCTTCACTTCCATCATGGATGCACTA.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |