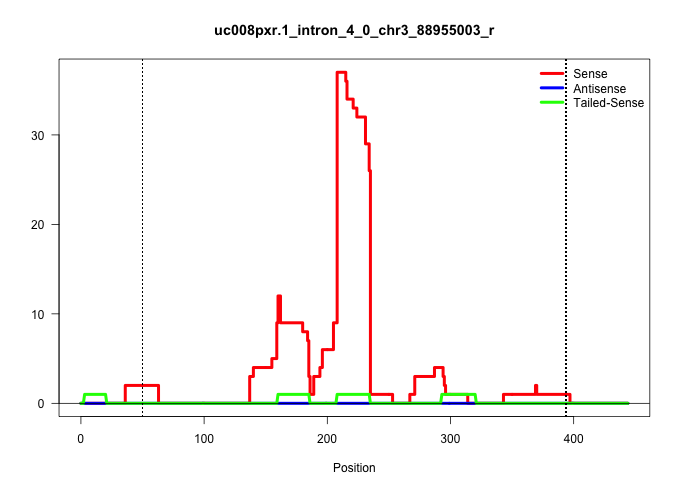
| Gene: Hcn3 | ID: uc008pxr.1_intron_4_0_chr3_88955003_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(6) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| TGCTGCAGGACTTCCCGTCCGACTGCTGGGTCTCCATGAACCGCATGGTGGTGAGAAGCCCCCCCAGCTCTGGTACTCCTGGGTCACAGGGAGCAGAAGGCAGGAGATGATCCAGAAGGAAAGCAGCAGGCAGGAGATGGGATTGCGGGGAGGAGAAGGGTAAGGAACGTAGATAGATGGAGGAAGTGTTTACATGTCAGAGGAATCAGACAAAGAAGGGAACCTAGGCAAGACAAGGGTCCCCACCAACGGAAGAAACCCAACAAATGCCTGCCGAGAGGAAGGTCTGAGGTAGAAGGGGTACGCACAGAACTTAAAGGGAGAAAGGGGGGCAGGTCGAGAATGAGACTCGGAGGGGCCCAAGCCCTGTTCTGCAACATCCTCTGCCCCCCAGAACCACTCGTGGGGCCGCCAGTATTCCCACGCCCTGTTCAAGGCCATGAG .............................................................................................................................................................................................................((((((.............((((((......((((..((......)).....))))......))))))((....))...)))))).........(((..........(((.....)))...((((((((((.((((.......))))..(((((....)))))..............))))))))))...))).............................................. ...........................................................................................................................................................................................................204..................................................................................................................................................................................................401......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................GACAAAGAAGGGAACCTAGGCAAGACA................................................................................................................................................................................................................. | 27 | 1 | 25.00 | 25.00 | 12.00 | 7.00 | 1.00 | - | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCAGACAAAGAAGGGAACCTAGGCAA..................................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GACAAAGAAGGGAACCTAGGCAAGAC.................................................................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................TGGGATTGCGGGGAGGAGAAGGGTA.......................................................................................................................................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................TGAACCGCATGGTGGTGAGAAGCCCCC............................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GTAAGGAACGTAGATAGATGGAGGAA................................................................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TAAGGAACGTAGATAGATGGAGGAA................................................................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................TGAGGTAGAAGGGGTACGCACAGAACT.................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TCAGAGGAATCAGACAAAGAAGGGAACC............................................................................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTACATGTCAGAGGAATCAGACAAAG..................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GACAAAGAAGGGAACCTAGGCAAGACt................................................................................................................................................................................................................. | 27 | t | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TAAGGAACGTAGATAGATGGAGGAAG.................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................TTCTGCAACATCCTCTGCCCCCCAGAAC............................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................GGAACGTAGATAGATGGAGGAAGTGTT.............................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GATTGCGGGGAGGAGAAGGGTAA......................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TACATGTCAGAGGAATCAGACAAAGA.................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGCAGGACTTCCCGTCtg....................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | tg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................................AGAAGGGGTACGCACAGAACTTAAtggt........................................................................................................................... | 28 | tggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................TGAGACTCGGAGGGGCCCAAGCCCTGT.......................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GTAAGGAACGTAGATAGATGGAGGAAG.................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GTAAGGAACGTAGATAGATGGAGGA.................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TAAGGAACGTAGATAGATGGAGGAAa.................................................................................................................................................................................................................................................................. | 26 | a | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTACATGTCAGAGGAATCAGACAAAGA.................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TGTCAGAGGAATCAGACAAAGAAGGGA............................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................TGCCGAGAGGAAGGTCTGAGGTA...................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................AAGGGTAAGGAACGTAGATAGATGG........................................................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................TGCCTGCCGAGAGGAAGGTCTGAGGTAG..................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................................................TAGGCAAGACAAGGGTCCCCACCAACGGA............................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................TGCCGAGAGGAAGGTCTGAGGTAGA.................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| TGCTGCAGGACTTCCCGTCCGACTGCTGGGTCTCCATGAACCGCATGGTGGTGAGAAGCCCCCCCAGCTCTGGTACTCCTGGGTCACAGGGAGCAGAAGGCAGGAGATGATCCAGAAGGAAAGCAGCAGGCAGGAGATGGGATTGCGGGGAGGAGAAGGGTAAGGAACGTAGATAGATGGAGGAAGTGTTTACATGTCAGAGGAATCAGACAAAGAAGGGAACCTAGGCAAGACAAGGGTCCCCACCAACGGAAGAAACCCAACAAATGCCTGCCGAGAGGAAGGTCTGAGGTAGAAGGGGTACGCACAGAACTTAAAGGGAGAAAGGGGGGCAGGTCGAGAATGAGACTCGGAGGGGCCCAAGCCCTGTTCTGCAACATCCTCTGCCCCCCAGAACCACTCGTGGGGCCGCCAGTATTCCCACGCCCTGTTCAAGGCCATGAG .............................................................................................................................................................................................................((((((.............((((((......((((..((......)).....))))......))))))((....))...)))))).........(((..........(((.....)))...((((((((((.((((.......))))..(((((....)))))..............))))))))))...))).............................................. ...........................................................................................................................................................................................................204..................................................................................................................................................................................................401......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................................................................................................................................GCCCAAGCCCTGTTCTtacc....................................................................... | 20 | tacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |