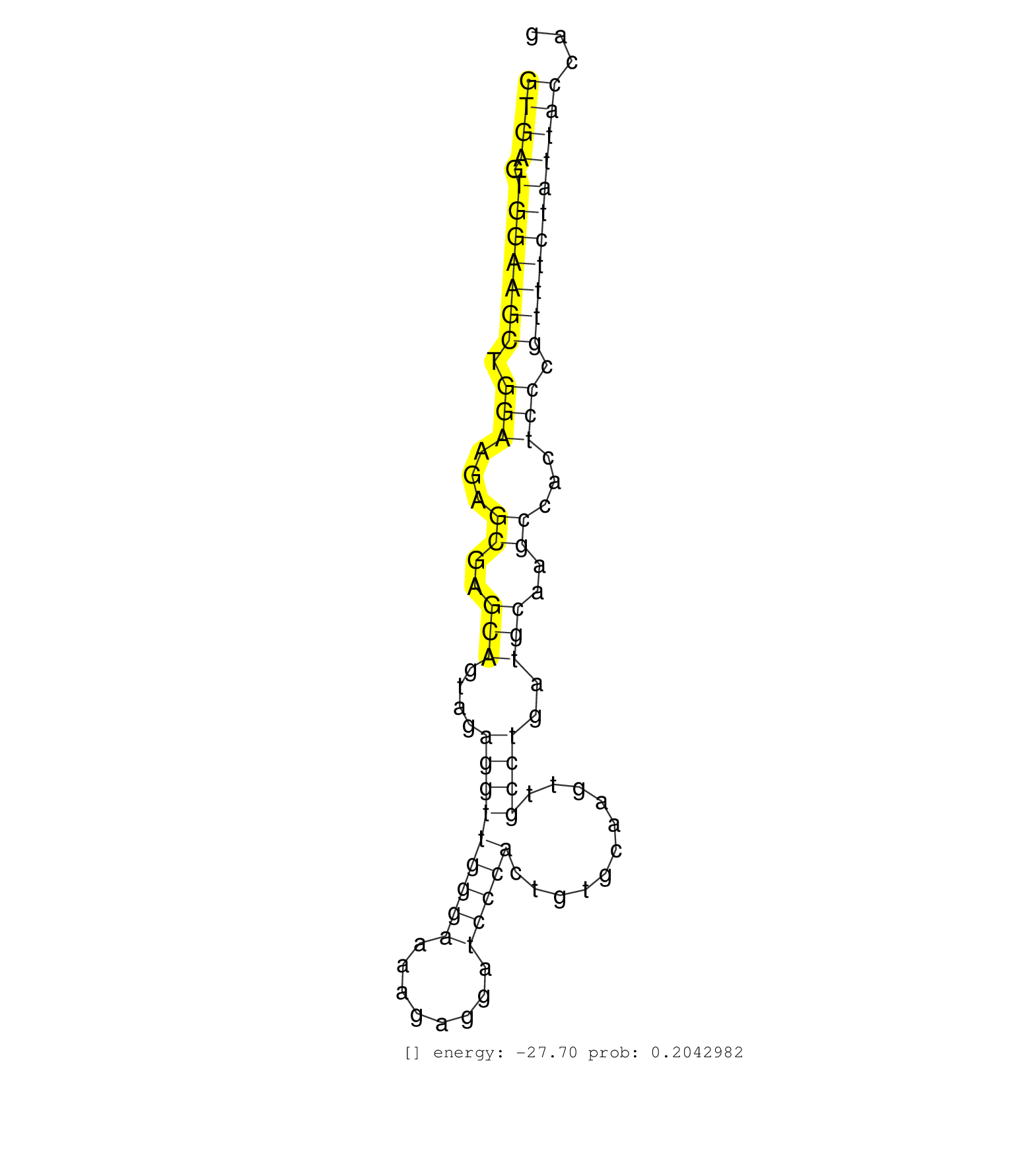

| Gene: Msto1 | ID: uc008pxd.1_intron_6_0_chr3_88715362_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(3) PIWI.mut |
(26) TESTES |
| GGCGGGGAGTCCTAACCTGGGGCCTGCTCCCGGGCCCCTACAGTCTTGGGGTGAGTGGAAGCTGGAAGAGCGAGCAGTAGAGGTTGGGAAAAGAGGATCCCACTGTGCAAGTTGCCTGATGCAAGCCACTCCCGTTTCTATTACCAGGAGCCTCAGAAAAACATCTATCGTCTCTTAAACACGGCATTTGGTCTGGT ..................................................((((.(((((((.(((...((..(((....(((((((((........)))))...........))))..)))..))...))).)))))))))))..................................................... ..................................................51..............................................................................................147................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGCA......................................................................................................................... | 26 | 1 | 70.00 | 70.00 | 31.00 | 11.00 | 12.00 | 3.00 | 1.00 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGC.......................................................................................................................... | 25 | 1 | 21.00 | 21.00 | 3.00 | 6.00 | 4.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGCAG........................................................................................................................ | 27 | 1 | 15.00 | 15.00 | - | 2.00 | 2.00 | 2.00 | 1.00 | 4.00 | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGCAGTA...................................................................................................................... | 29 | 1 | 7.00 | 7.00 | 5.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGCAGT....................................................................................................................... | 28 | 1 | 6.00 | 6.00 | 2.00 | - | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGGAAGCTGGAAGAGCGAGCA......................................................................................................................... | 25 | 1 | 5.00 | 5.00 | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGCAt........................................................................................................................ | 27 | t | 5.00 | 70.00 | 2.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGGAAGCTGGAAGAGCGAGCAG........................................................................................................................ | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGCAa........................................................................................................................ | 27 | a | 3.00 | 70.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TATCGTCTCTTAAACACGGCA........... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................AAAAACATCTATCGTCTCTTAAACACGGC............ | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAG........................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................GAAAAACATCTATCGTCTCTTAAACA................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGggca......................................................................................................................... | 26 | ggca | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGCTGGAAGAGCGAGCAGTAGAGGTTG............................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGaag........................................................................................................................ | 27 | aag | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGcgcc......................................................................................................................... | 26 | cgcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................AGTGGAAGCTGGAAGAGCGAGCAGTA...................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGCTGGAAGAGCGAGCAGTAGAGGT................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AACATCTATCGTCTCTTAAACACGGCA........... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................AGAAAAACATCTATCGTCTCTTAAACACGG............. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGCt......................................................................................................................... | 26 | t | 1.00 | 21.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGA............................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TCGTCTCTTAAACACGGCATTTGGTCTG.. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGga......................................................................................................................... | 26 | ga | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TCAGAAAAACATCTATCGTCTCTTAAA.................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AACATCTATCGTCTCTTAAACACGGCAT.......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................AACATCTATCGTCTCTTAAACACGG............. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................ATCGTCTCTTAAACACGGCA........... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGCc......................................................................................................................... | 26 | c | 1.00 | 21.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TCTATCGTCTCTTAAACACGGCATTTGGTC.... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGGAAGCTGGAAGAGCGAGCAt........................................................................................................................ | 26 | t | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGGAAGCTGGAAGAGCGAGCAaa....................................................................................................................... | 28 | aa | 1.00 | 70.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GAAAAACATCTATCGTCTCTTAAACACGGC............ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................GAGTGGAAGCTGGAAGAGCGAGCAGTA...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGGAAGCTGGAAGAG............................................................................................................................... | 16 | 19 | 0.05 | 0.05 | - | - | - | - | - | - | - | - | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGCGGGGAGTCCTAACCTGGGGCCTGCTCCCGGGCCCCTACAGTCTTGGGGTGAGTGGAAGCTGGAAGAGCGAGCAGTAGAGGTTGGGAAAAGAGGATCCCACTGTGCAAGTTGCCTGATGCAAGCCACTCCCGTTTCTATTACCAGGAGCCTCAGAAAAACATCTATCGTCTCTTAAACACGGCATTTGGTCTGGT ..................................................((((.(((((((.(((...((..(((....(((((((((........)))))...........))))..)))..))...))).)))))))))))..................................................... ..................................................51..............................................................................................147................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|