
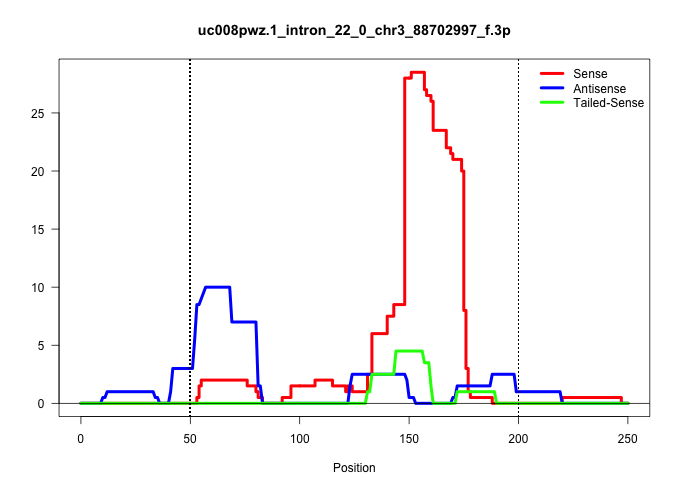
| Gene: Gon4l | ID: uc008pwz.1_intron_22_0_chr3_88702997_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(5) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(28) TESTES |
| GTTAAAGGCTGTCCAGTTGCCCCTCCGAGGAAAGGAGTTGTGCCTTGCTGTTTGTTCTGTGTGTTGTTACTCCTTGGGTCAGAGGAGTGCCTGTGAAGGGAGCTCTCAGAGCCGTGTGAGCACAGCTGCCTTCTGTGCCCTTGTCGGTTAGGATTGCAGGCCCACAGAAGGATGCTGACCAGTGCAAGTATTCCTTGTAGGTACGTGAAGCTCTCCAGCACACCCCTGGCAAATACGAAGATTTCCTTCA .....................................................((((((...(((..((((((((......))))))))(((....))).)))...))))))..........((((..(((((((((..((((.((((....))))))))..)))))))))..))))......................................................................... ..................................................51..............................................................................................................................179..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................TAGGATTGCAGGCCCACAGAAGGATGC........................................................................... | 27 | 2 | 12.00 | 12.00 | 5.00 | 3.50 | - | 1.00 | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ....................................................................................................................................................TAGGATTGCAGGCCCACAGAAGGATGCT.......................................................................... | 28 | 2 | 4.50 | 4.50 | 1.00 | 1.00 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - |
| .....................................................................................................................................TGTGCCCTTGTCGGTTAGGATTGCAGGC......................................................................................... | 28 | 2 | 3.00 | 3.00 | - | - | 1.00 | - | 0.50 | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ............................................................................................................................................TTGTCGGTTAGGATTGCAGGCCCACAG................................................................................... | 27 | 2 | 1.50 | 1.50 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| ....................................................................................................................................................TAGGATTGCAGGCCCACAGAAGGATGCTG......................................................................... | 29 | 2 | 1.50 | 1.50 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................CGGTTAGGATTGCtggt......................................................................................... | 17 | tggt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAGGATTGCAGGCCCACAGAAGGATG............................................................................ | 26 | 2 | 1.00 | 1.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TCTGTGCCCTTGTCGGTTAGGATTGC............................................................................................. | 26 | 2 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TCTGTGCCCTTGTCGGTTAGGATTtc............................................................................................. | 26 | tc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGCTGTTTGTTCTGTGTGTTGTTACTC.................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGTGCCCTTGTCGGTTAGGATTGC............................................................................................. | 24 | 2 | 1.00 | 1.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ............................................................................................................................................................................TGCTGACCAGTGCAgacg............................................................ | 18 | gacg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GTACGTGAAGCTCTCgct................................ | 18 | gct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGTGCCCTTGTCGGTTAGGATTGgagg.......................................................................................... | 27 | gagg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................CGGTTAGGATTGCtgg.......................................................................................... | 16 | tgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCGGTTAGGATTGCAGGCCCACAGAAG................................................................................ | 27 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TCTGTGTGTTGTTACTCCTTGGGTCAGA....................................................................................................................................................................... | 28 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................CCACAGAAGGATGCTGACCAGTGCAAG.............................................................. | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ...................................................................................................................................TCTGTGCCCTTGTCGGTTAGGATTGCA............................................................................................ | 27 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGTGCCCTTGTCGGTTAGGATTGCAGG.......................................................................................... | 27 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TTCTGTGTGTTGTTACTCCTTGGGTC.......................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................CAGCTGCCTTCTGTGCCCTTGTCGGTT..................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................AGGGAGCTCTCAGAGCCGTGTGAGCACA.............................................................................................................................. | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCGGTTAGGATTGCAGGCCCACAGAA................................................................................. | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................AGGATTGCAGGCCCACAGAAGGATGCTG......................................................................... | 28 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CACCCCTGGCAAATACGAAGATTTCCT... | 27 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................GATTGCAGGCCCACAGAAGGATGCT.......................................................................... | 25 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAGGATTGCAGGCCCACAGAAGGATGCTGA........................................................................ | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| .....................................................GTTCTGTGTGTTGTTACTCCTTG.............................................................................................................................................................................. | 23 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGTGCCCTTGTCGGTTAGGATTGCAGGt......................................................................................... | 28 | t | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................AGGGAGCTCTCAGAGCCGTGTGAGC................................................................................................................................. | 25 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................AGAGCCGTGTGAGCACAGCTGCCTTC..................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GTGAAGGGAGCTCTCAGAGCCGT....................................................................................................................................... | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TTCTGTGTGTTGTTACTCCTTGGGTCA......................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTTAAAGGCTGTCCAGTTGCCCCTCCGAGGAAAGGAGTTGTGCCTTGCTGTTTGTTCTGTGTGTTGTTACTCCTTGGGTCAGAGGAGTGCCTGTGAAGGGAGCTCTCAGAGCCGTGTGAGCACAGCTGCCTTCTGTGCCCTTGTCGGTTAGGATTGCAGGCCCACAGAAGGATGCTGACCAGTGCAAGTATTCCTTGTAGGTACGTGAAGCTCTCCAGCACACCCCTGGCAAATACGAAGATTTCCTTCA .....................................................((((((...(((..((((((((......))))))))(((....))).)))...))))))..........((((..(((((((((..((((.((((....))))))))..)))))))))..))))......................................................................... ..................................................51..............................................................................................................................179..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................GTTCTGTGTGTTGTTACTCCTTGGGTCA......................................................................................................................................................................... | 28 | 2 | 3.00 | 3.00 | 0.50 | 1.00 | - | 0.50 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TGTTCTGTGTGTTGTTACTCCTTGGGTCA......................................................................................................................................................................... | 29 | 2 | 2.50 | 2.50 | 0.50 | 1.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................CCTTGCTGTTTGTTCTGTGTGTTGTTA..................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGCTGACCAGTGCAAGTATTCCTTGTA................................................... | 27 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TTCTGTGTGTTGTTACTCCTTGGGTCAaa......................................................................................................................................................................... | 29 | aa | 1.00 | 0.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AGCTGCCTTCTGTGCCCTTGTCGGTTA.................................................................................................... | 27 | 2 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................GCCTTGCTGTTTGTTCTGTGTGTTGTTA..................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TATTCCTTGTAGGTACGTGAAGCTCTCCAGCA.............................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................CTGTGTGTTGTTACTCCTTGGGTCAGA....................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GCTGCCTTCTGTGCCCTTGTCGGTTA.................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........GTCCAGTTGCCCCTCCGAGGAAAGGA...................................................................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GCTGCCTTCTGTGCCCTTGTCGGTTAGGA................................................................................................. | 29 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AGCTGCCTTCTGTGCCCTTGTCGGTT..................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................GATGCTGACCAGTGCAAGTATTCCTTGTA................................................... | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CCAGTTGCCCCTCCGAGGAAAG........................................................................................................................................................................................................................ | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| .........................................................TGTGTGTTGTTACTCCTTGGGTCAGA....................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .......................................................TCTGTGTGTTGTTACTCCTTGGGTCAGA....................................................................................................................................................................... | 28 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |