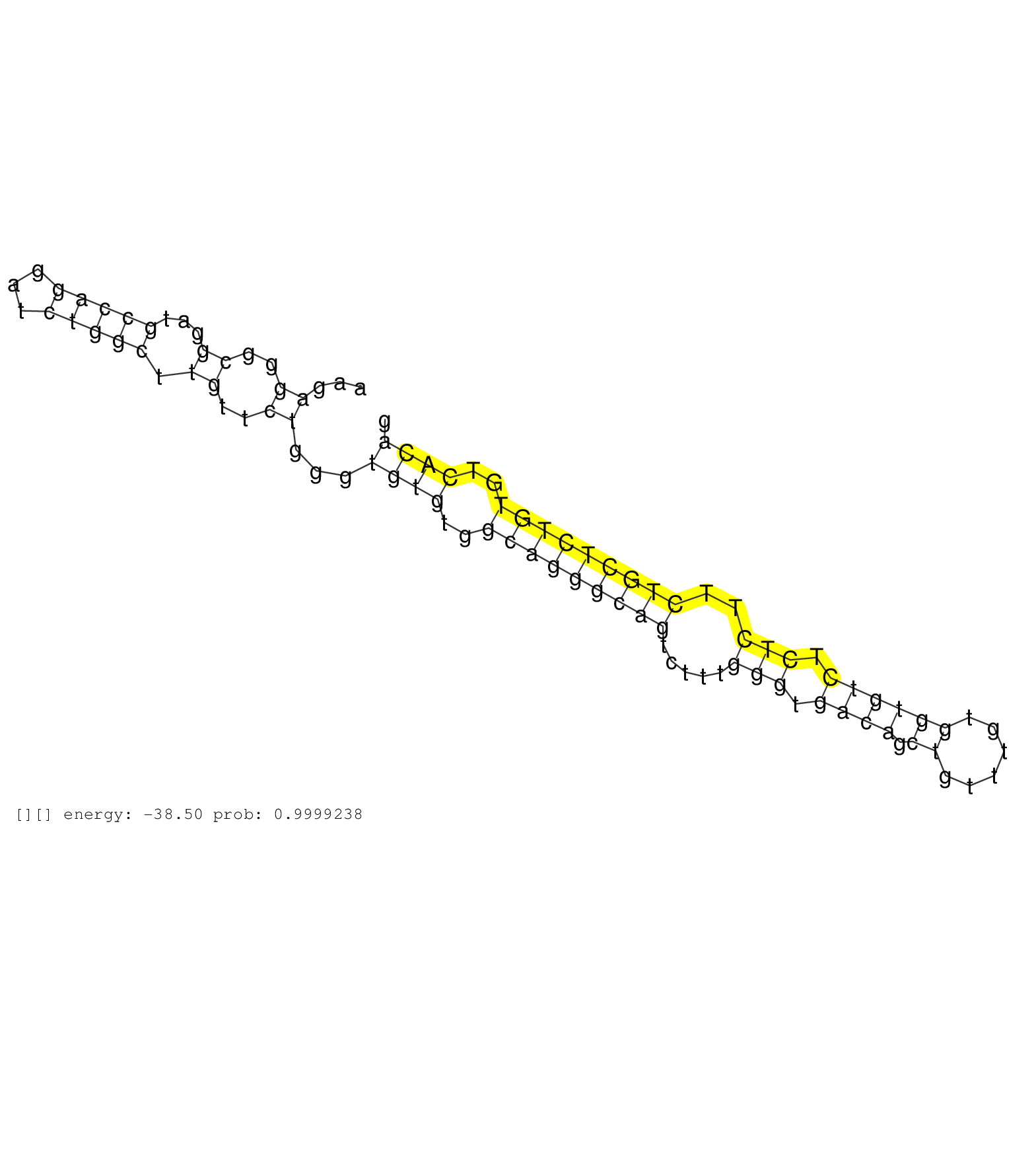

| Gene: Arhgef2 | ID: uc008pvu.1_intron_3_0_chr3_88435901_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(10) TESTES |
| ACAACCGCTGTAAAGACACCCTGGCCAACTGTACCAAGGTCAAGCAGAAGGTGAGCTAGATGGCAGGGTGGGCAGAGGGCTTGCGGGGAGGGCATCGCCATCCCTGCGGGCCTTTCCACACATCCCTCACCCTCTGTCCCTCTCCTTGCCGTGGGGCATCCCAGACCCTCGGCCTGTGTAAACACAGCCCTGCCACACTAGTTTACCTATATCCCGATGTCCTTAACTGCGGGGTCAACTTTCTCCCAAGAGGGCGGATGCCAGGATCTGGCTTGTTCTGGGTGTGTGGCAGGGCAGTCTTTGGGTGACAGCTGTTTGTGGTGTCTCTCTTCTGCTCTGTGTCACAGCAACAGAAAGCTGCACTGCTGAGGAACAACACTGCTTTGCAGTCTGTCTC ..........................................................................................................................................................................................................................................................((..((...(((((...))))).))..))...((((..(((((((((.....(((.((((.((......)))))).)))..)))))))))..))))................................................... .......................................................................................................................................................................................................................................................248................................................................................................347................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................CAACTGTACCAAGGTCAAGCAG.............................................................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - |
| .....CGCTGTAAAGACACCCTGGCCAACT............................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................ACAGAAAGCTGCACTGCTGAGGAACA...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............GACACCCTGGCCAACTGTACCAAGGTCAAGCA............................................................................................................................................................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................CTCTCTTCTGCTCTGTGTCcc.................................................... | 21 | cc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGAGCTAGATGGCAGGGTGGGCAGA................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................ACAGAAAGCTGCACTGCTGAGGAACAAC.................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................................TCTCTTCTGCTCTGTGTCA..................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................CAACTGTACCAAGGTCAAGCA............................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................................................................................................................................................................TCTCTTCTGCTCTGTGTCACAGt................................................. | 23 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ACAACCGCTGTAAAGACACCCTGGCCAACTGTACCAAGGTCAAGCAGAAGGTGAGCTAGATGGCAGGGTGGGCAGAGGGCTTGCGGGGAGGGCATCGCCATCCCTGCGGGCCTTTCCACACATCCCTCACCCTCTGTCCCTCTCCTTGCCGTGGGGCATCCCAGACCCTCGGCCTGTGTAAACACAGCCCTGCCACACTAGTTTACCTATATCCCGATGTCCTTAACTGCGGGGTCAACTTTCTCCCAAGAGGGCGGATGCCAGGATCTGGCTTGTTCTGGGTGTGTGGCAGGGCAGTCTTTGGGTGACAGCTGTTTGTGGTGTCTCTCTTCTGCTCTGTGTCACAGCAACAGAAAGCTGCACTGCTGAGGAACAACACTGCTTTGCAGTCTGTCTC ..........................................................................................................................................................................................................................................................((..((...(((((...))))).))..))...((((..(((((((((.....(((.((((.((......)))))).)))..)))))))))..))))................................................... .......................................................................................................................................................................................................................................................248................................................................................................347................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................AGACCCTCGGCCTGtggc............................................................................................................................................................................................................................. | 18 | tggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................................TACCTATATCCCGATGTCCTTAct............................................................................................................................................................................ | 24 | ct | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - |