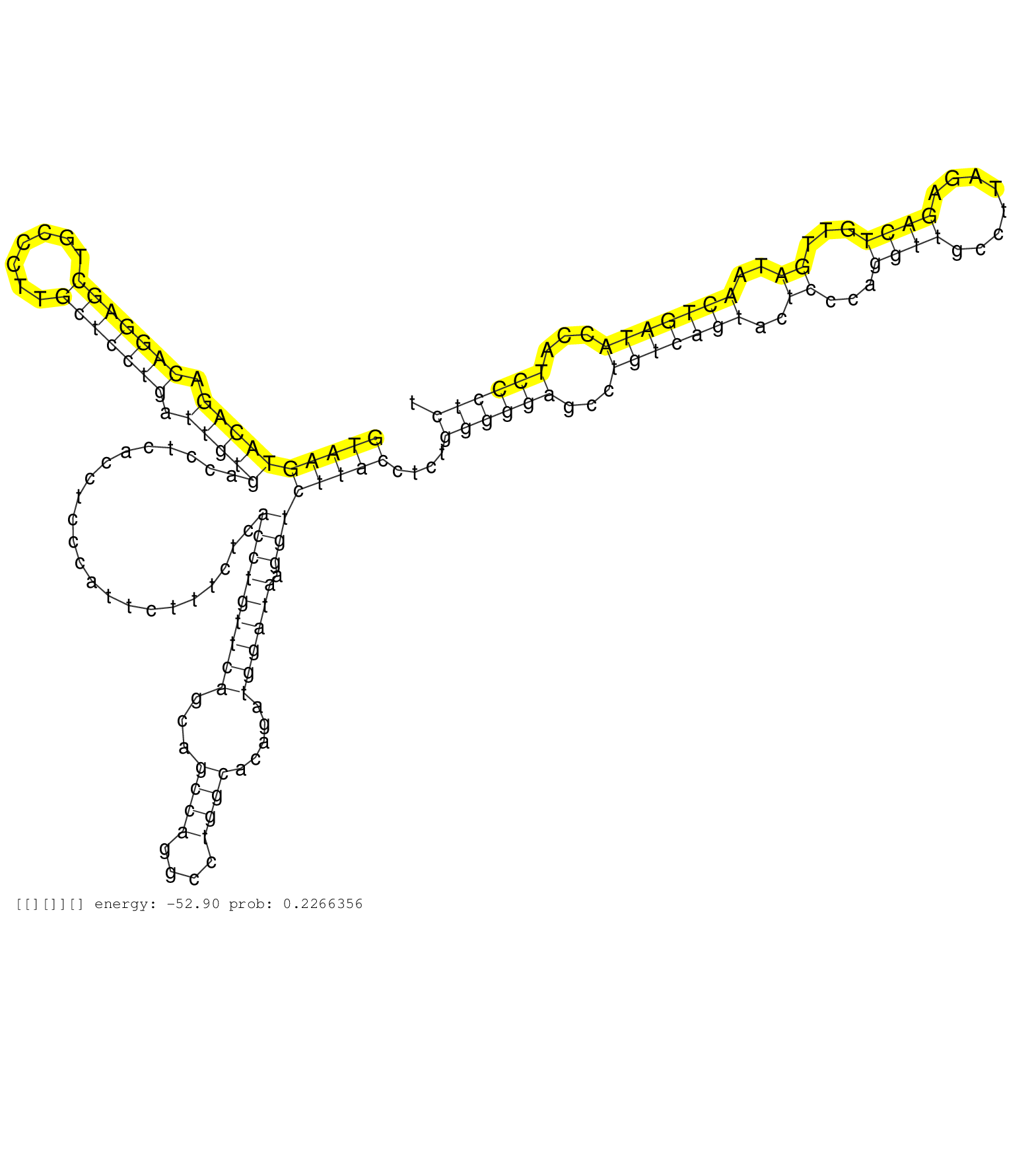
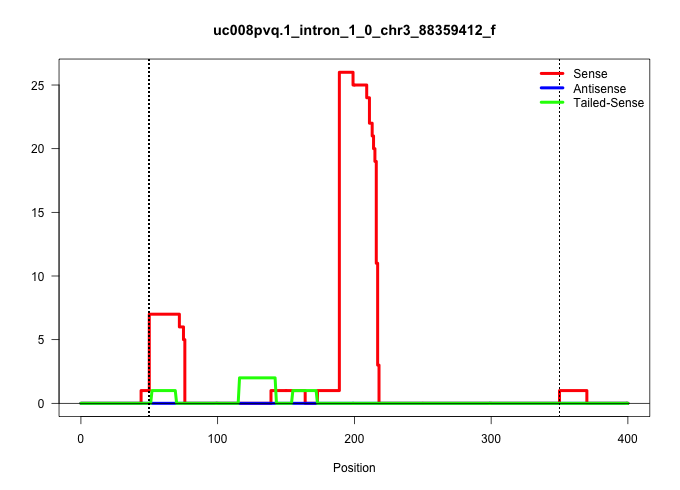
| Gene: Ubqln4 | ID: uc008pvq.1_intron_1_0_chr3_88359412_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(3) PIWI.mut |
(15) TESTES |
| GGCATCAAGGATGGGCTCACAGTCCACCTAGTCATCAAAACCCCTCAGAAGTAAGTACAGACAGGAGCTGCCCTTGCTCCTGATTGTGACCTCACCTCCCATTCTTTCTCACCTGTTCAGCAGCCAGGCCTGGCACAGATGGATAAGGTCTTACCTCTGGGGGAGCCTGTCAGTACTCCCAGGTTGCCTTAGAGACTGTTGATAACTGATACCATCCCTCTCCCCAGCATCCTGCTAGCCTGGGGTAAGGGTGGAACAAGGCTGCTTAGGAGGTGGCTTGAGCTGTCAGTCCAAGGCCACCTTGTCACTTGTGCACACTCCAGCCAGGTTTCCCTACTCTTTGTCCTCAGGGCTCAAGATCCAGTGACTGCTGCTGCTTCTCCTCCATCCACTCCTGACT ..................................................((((((((((.(((((((.......))))))).)))))......................(((((((((...((((....)))).....)))))).))))))))....((((((...(((((((..((...((((........))))...))..)))))))...)))))).................................................................................................................................................................................... ..................................................51........................................................................................................................................................................221................................................................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................TAGAGACTGTTGATAACTGATACCATC........................................................................................................................................................................................ | 27 | 1 | 10.00 | 10.00 | 5.00 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................TAGAGACTGTTGATAACTGATACCATCC....................................................................................................................................................................................... | 28 | 1 | 8.00 | 8.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTAAGTACAGACAGGAGCTGCCCTTG.................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 |
| .............................................................................................................................................................................................TAGAGACTGTTGATAACTGATACCATCCC...................................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TAGAGACTGTTGATAACTGATA............................................................................................................................................................................................. | 22 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCAGCAGCCAGGCCTGGCACAGATGGt................................................................................................................................................................................................................................................................. | 27 | t | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AAGTACAGACAGGAtccc.......................................................................................................................................................................................................................................................................................................................................... | 18 | tccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................................................................................................TGGGGTAAGGGTGGAACA.............................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................GGCTCAAGATCCAGTGACTG.............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGGATAAGGTCTTACCTCTGGGGGA............................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TAGAGACTGTTGATAACTGATACCA.......................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TAGAGACTGTTGATAACTGATACC........................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TCAGAAGTAAGTACAGACAGGAGCTGCC........................................................................................................................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAAGTACAGACAGGAGCTGCCCTT..................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TAGAGACTGTTGATAACTGATACCAT......................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TACTCCCAGGTTGCCTTAGAGACTGT......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................CTCTGGGGGAGCCTGagaa................................................................................................................................................................................................................................... | 19 | agaa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TAGAGACTGTTGATAACTGA............................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TCTGGGGGAGCCTGaaaa................................................................................................................................................................................................................................... | 18 | aaaa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| GGCATCAAGGATGGGCTCACAGTCCACCTAGTCATCAAAACCCCTCAGAAGTAAGTACAGACAGGAGCTGCCCTTGCTCCTGATTGTGACCTCACCTCCCATTCTTTCTCACCTGTTCAGCAGCCAGGCCTGGCACAGATGGATAAGGTCTTACCTCTGGGGGAGCCTGTCAGTACTCCCAGGTTGCCTTAGAGACTGTTGATAACTGATACCATCCCTCTCCCCAGCATCCTGCTAGCCTGGGGTAAGGGTGGAACAAGGCTGCTTAGGAGGTGGCTTGAGCTGTCAGTCCAAGGCCACCTTGTCACTTGTGCACACTCCAGCCAGGTTTCCCTACTCTTTGTCCTCAGGGCTCAAGATCCAGTGACTGCTGCTGCTTCTCCTCCATCCACTCCTGACT ..................................................((((((((((.(((((((.......))))))).)))))......................(((((((((...((((....)))).....)))))).))))))))....((((((...(((((((..((...((((........))))...))..)))))))...)))))).................................................................................................................................................................................... ..................................................51........................................................................................................................................................................221................................................................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|