
| Gene: Mapbpip | ID: uc008pvo.1_intron_1_0_chr3_88354708_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(12) TESTES |
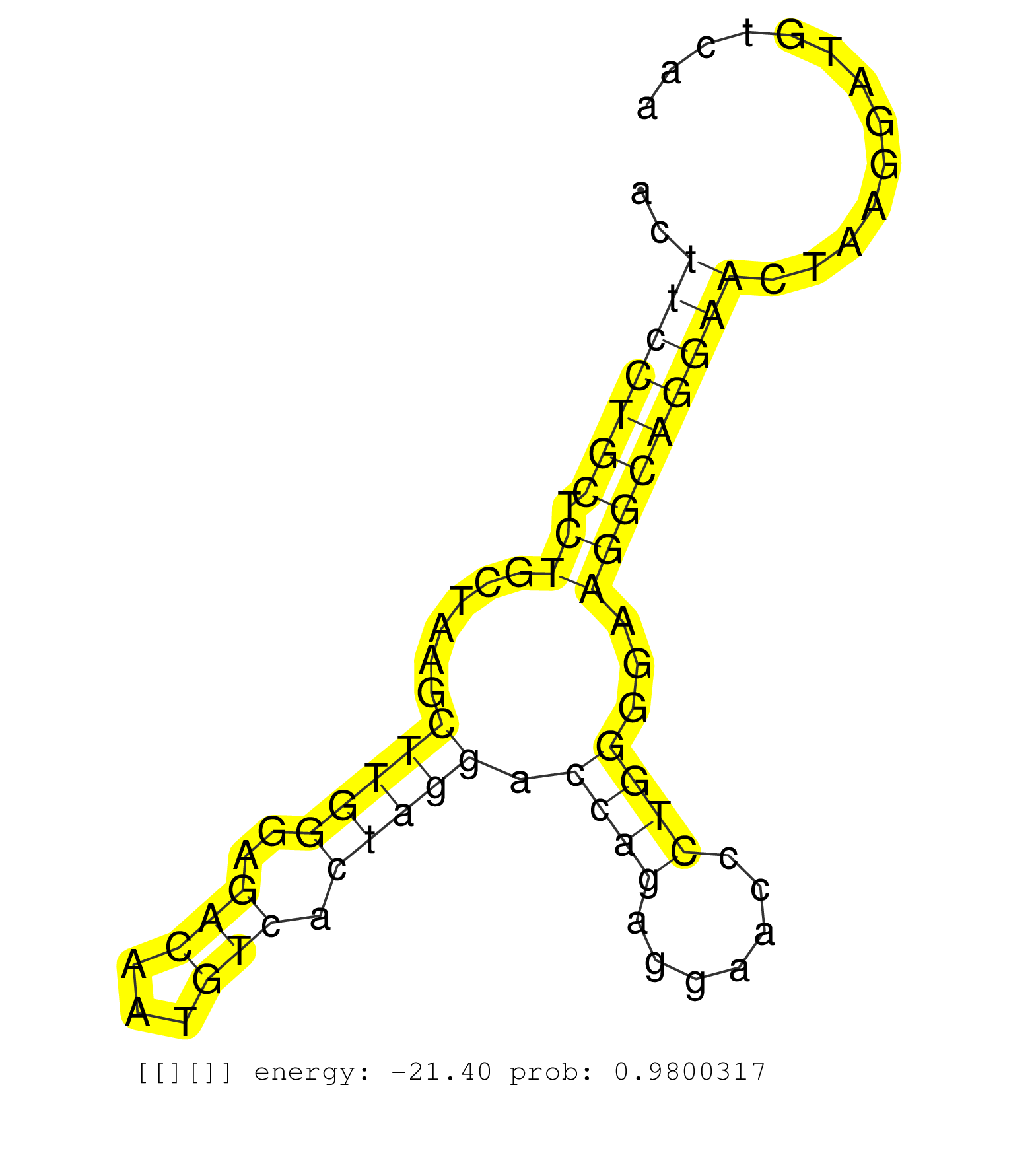
| CTTGGTTAGCACATGGCTCAGCTGAGGCTGGAATCCAGTTTCAGACCCCCATGTCTTAAATGCTGGGCAGAGCTCCTAGCTGAACTTCCTGCTCTGCTAAGCTTGGGAGACAATGTCACTAGGACCAGAGGAACCCTGGGGAAGGCAGGAACTAAGGATGTCAAGCAGAATGCCCCAAAGTATCATGTCCTTCCCACCAGGAGGGCCGTGTAGCCATTACGAGGGTGGCCAACCTTCTGCTATGTATGTA .....................................................................................(((((((.((......(((((..(((...))).))))).((((.......))))...)))))))))................................................................................................... ...................................................................................84..............................................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......TAGCACATGGCTCAGCTGAGGCTGG........................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................CTGCTCTGCTAAGCTTGGGAGACAATGT...................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................CTGGGGAAGGCAGGAACTAAGGATG.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CCTGGGGAAGGCAGGAACTAAGGAT........................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............TGGCTCAGCTGAGGCTGctct........................................................................................................................................................................................................................ | 21 | ctct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................TGGGGAAGGCAGGAACTAAGGA............................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................GGAACTAAGGATGTCAAGCAGAATGCCC........................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................................................................AGGGCCGTGTAGCCATTACGAGG.......................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................TGCTAAGCTTGGGAGACAATGTCACT.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| CTTGGTTAGCACATGGCTCAGCTGAGGCTGGAATCCAGTTTCAGACCCCCATGTCTTAAATGCTGGGCAGAGCTCCTAGCTGAACTTCCTGCTCTGCTAAGCTTGGGAGACAATGTCACTAGGACCAGAGGAACCCTGGGGAAGGCAGGAACTAAGGATGTCAAGCAGAATGCCCCAAAGTATCATGTCCTTCCCACCAGGAGGGCCGTGTAGCCATTACGAGGGTGGCCAACCTTCTGCTATGTATGTA .....................................................................................(((((((.((......(((((..(((...))).))))).((((.......))))...)))))))))................................................................................................... ...................................................................................84..............................................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................CCATGTCTTAAATGCTGGGCAGAGCTCCT............................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................CCAAAGTATCATGTCCTTCCCACCAGGA................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................GGGAGACAATGTCACTAGGACCAGA......................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AGTATCATGTCCTTCCC....................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 |