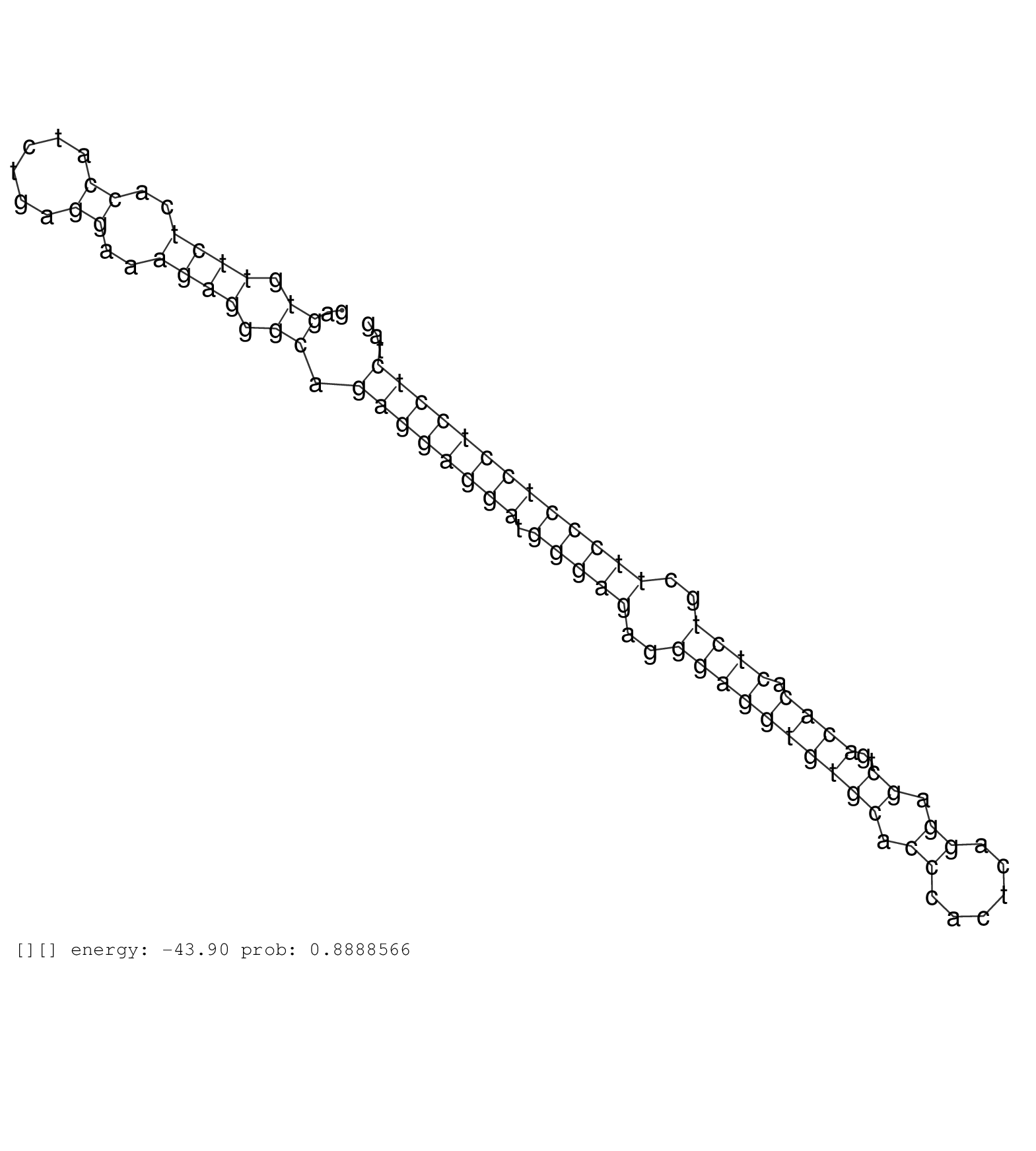
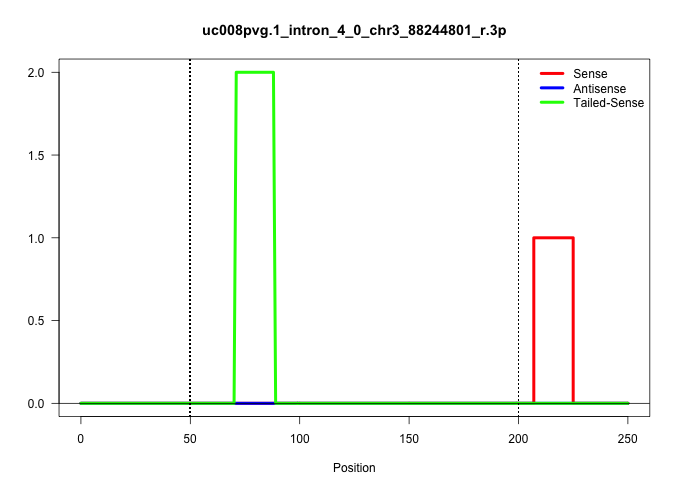
| Gene: Sema4a | ID: uc008pvg.1_intron_4_0_chr3_88244801_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(6) TESTES |
| GTTCCGCCCCGCCCCGCCCCGCCCCGCCCCGCCCCGCGCTGGGTATTTTTCATGAATTAGGCATTTTCAACCTCACAGAGAGCCTAGGAGCTCTGGGTCTGAGTGTTCTCACCATCTGAGGAAAGAGGGCAGAGGAGGATGGGAGAGGGAGGTGTGCACCCACTCAGGAGCTGACACACTCTGCTTCCCTCCTCCTCTAGTGCTCCATGGGCCCCTCCTCTGACAAAGCCTTGACCTTCATGAAGGACCA ......................................................................................................((.((((..((......))..)))).)).((((((((.(((((..((((((((((.((......)).))..)))).))))..)))))))))))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................TCTGAGGAAAGAGGGCAGAGGAGG................................................................................................................ | 24 | 1 | 7.00 | 7.00 | 6.00 | - | - | - | 1.00 | - |
| .......................................................................CTCACAGAGAGCCTtcac................................................................................................................................................................. | 18 | tcac | 2.00 | 0.00 | - | 2.00 | - | - | - | - |
| ...............................................................................................................................................................................................................TGGGCCCCTCCTCTGACA......................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| ..................................................................................................................TCTGAGGAAAGAGGGCAGAGGcgg................................................................................................................ | 24 | cgg | 1.00 | 0.00 | 1.00 | - | - | - | - | - |
| ................................................................................AGCCTAGGAGCTCTGGG......................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | 0.33 |
| GTTCCGCCCCGCCCCGCCCCGCCCCGCCCCGCCCCGCGCTGGGTATTTTTCATGAATTAGGCATTTTCAACCTCACAGAGAGCCTAGGAGCTCTGGGTCTGAGTGTTCTCACCATCTGAGGAAAGAGGGCAGAGGAGGATGGGAGAGGGAGGTGTGCACCCACTCAGGAGCTGACACACTCTGCTTCCCTCCTCCTCTAGTGCTCCATGGGCCCCTCCTCTGACAAAGCCTTGACCTTCATGAAGGACCA ......................................................................................................((.((((..((......))..)))).)).((((((((.(((((..((((((((((.((......)).))..)))).))))..)))))))))))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................AGCTCTGGGTCTGAGTGTTCTCACCATCTGA................................................................................................................................... | 31 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - |
| ....................................................................................GAGCTCTGGGTCTGAGaag................................................................................................................................................... | 19 | aag | 1.00 | 0.00 | - | - | - | 1.00 | - | - |