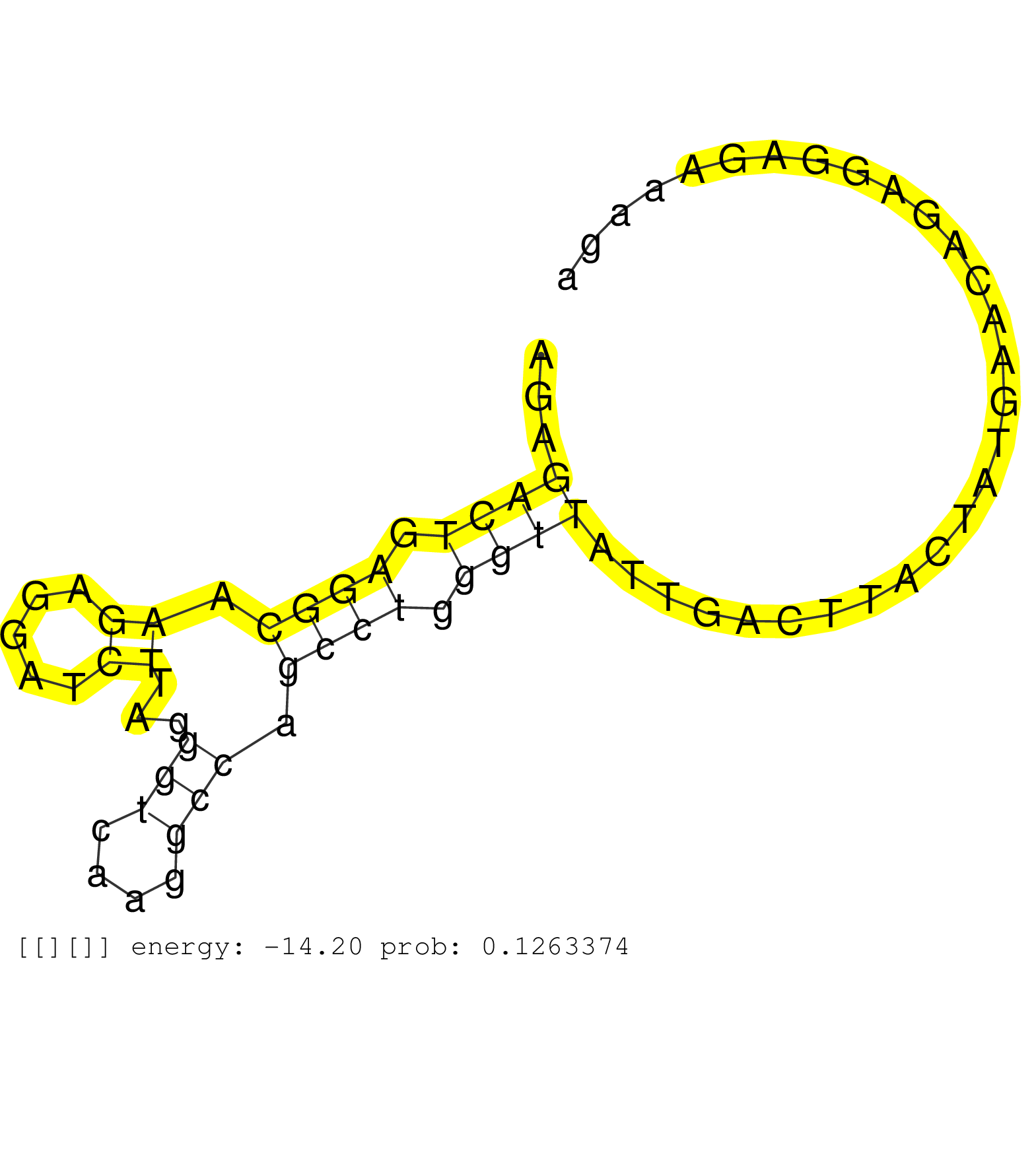
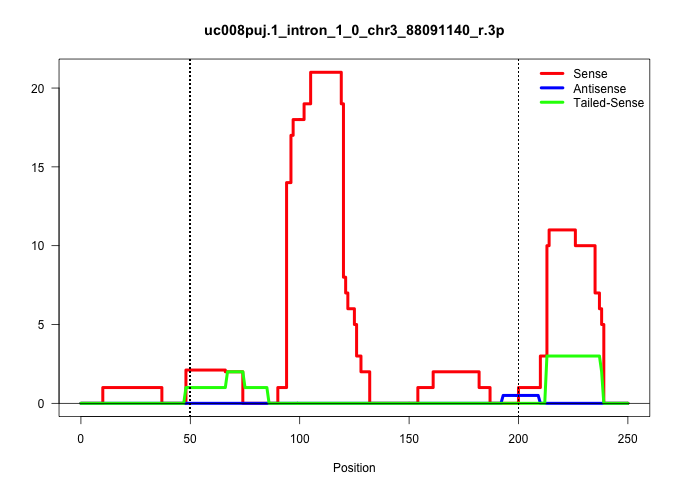
| Gene: 1700021C14Rik | ID: uc008puj.1_intron_1_0_chr3_88091140_r.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(19) TESTES |
| TGCAGGGATTTAATTTGAAAGACAGTCTTATTCCTGCAATCCCACCTTTGAGAGACTGAGGCAAGAGGATCTTAGGGTCAAGGCCAGCCTGGGTTATTGACTTACTATGAACAGAGGAGAAAGAGCAAGAAAAATAGGGTAGCGGAAAGCAAGCTAACCCTTGTTAGAATGAGTTTTGATGTAGTCACCTGGTTTTTCAGCCAAAGAGGAAGATAGCGTTGCGTGTTTTTGTCCAGCAAAATCCTCCCCC .....................................................((((.((((.((.....))...(((....))).)))).))))........................................................................................................................................................... ..................................................51.......................................................................124............................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................TATTGACTTACTATGAACAGAGGAGA.................................................................................................................................. | 26 | 1 | 11.00 | 11.00 | 3.00 | - | - | - | 3.00 | 1.00 | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TAGCGTTGCGTGTTTTTGTCCAGCAA........... | 26 | 1 | 4.00 | 4.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................TAGCGTTGCGTGTTTTTGTCCA............... | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................TGAGAGACTGAGGCAAGAGGATCTTA................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TAGCGTTGCGTGTTTTTGTCCAGCAt........... | 26 | t | 2.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TATGAACAGAGGAGAAAGAGCAAGAAA...................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GGGTTATTGACTTACTATGAACAGAGGAG................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TACTATGAACAGAGGAGAAAGAGCAA.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GATCTTAGGGTCAAGcgca.................................................................................................................................................................... | 19 | cgca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................................................TAGCGTTGCGTGTTTTTGTCCAGCt............ | 25 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TAGCGTTGCGTGTTTTTGTCCAGCA............ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AGATAGCGTTGCGTGTTTTTGTCCAGC............. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AGATAGCGTTGCGTGTTTTTGTCCA............... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TGTTAGAATGAGTTTTGATGTAGTCA............................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................AGCGTTGCGTGTTTTTGTCCAGCAA........... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TAACCCTTGTTAGAATGAGTTTTGATGT.................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........TAATTTGAAAGACAGTCTTATTCCTGC..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGACTTACTATGAACAGAGGAGAAAGAGC............................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TATTGACTTACTATGAACAGAGGAGAA................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TTGACTTACTATGAACAGAGGAGAAA................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................CCAAAGAGGAAGATAGCGTTGCGTGT........................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TTGACTTACTATGAACAGAGGAGAAAGAG............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TATTGACTTACTATGAACAGAGGAG................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TTGACTTACTATGAACAGAGGAGAAAGAGC............................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................TGAGAGACTGAGGCAAGAGGATCTTAa............................................................................................................................................................................... | 27 | a | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................TGAGAGACTGAGGCAAGA........................................................................................................................................................................................ | 18 | 9 | 0.11 | 0.11 | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGCAGGGATTTAATTTGAAAGACAGTCTTATTCCTGCAATCCCACCTTTGAGAGACTGAGGCAAGAGGATCTTAGGGTCAAGGCCAGCCTGGGTTATTGACTTACTATGAACAGAGGAGAAAGAGCAAGAAAAATAGGGTAGCGGAAAGCAAGCTAACCCTTGTTAGAATGAGTTTTGATGTAGTCACCTGGTTTTTCAGCCAAAGAGGAAGATAGCGTTGCGTGTTTTTGTCCAGCAAAATCCTCCCCC .....................................................((((.((((.((.....))...(((....))).)))).))))........................................................................................................................................................... ..................................................51.......................................................................124............................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................GTTTTTCAGCCAAAGaggg............................................ | 19 | aggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................TTCCTGCAATCCCAaaaa.............................................................................................................................................................................................................. | 18 | aaaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TCCAGCAAAATCCTCgatt.... | 19 | gatt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TTTTCAGCCAAAGAGGA........................................ | 17 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |