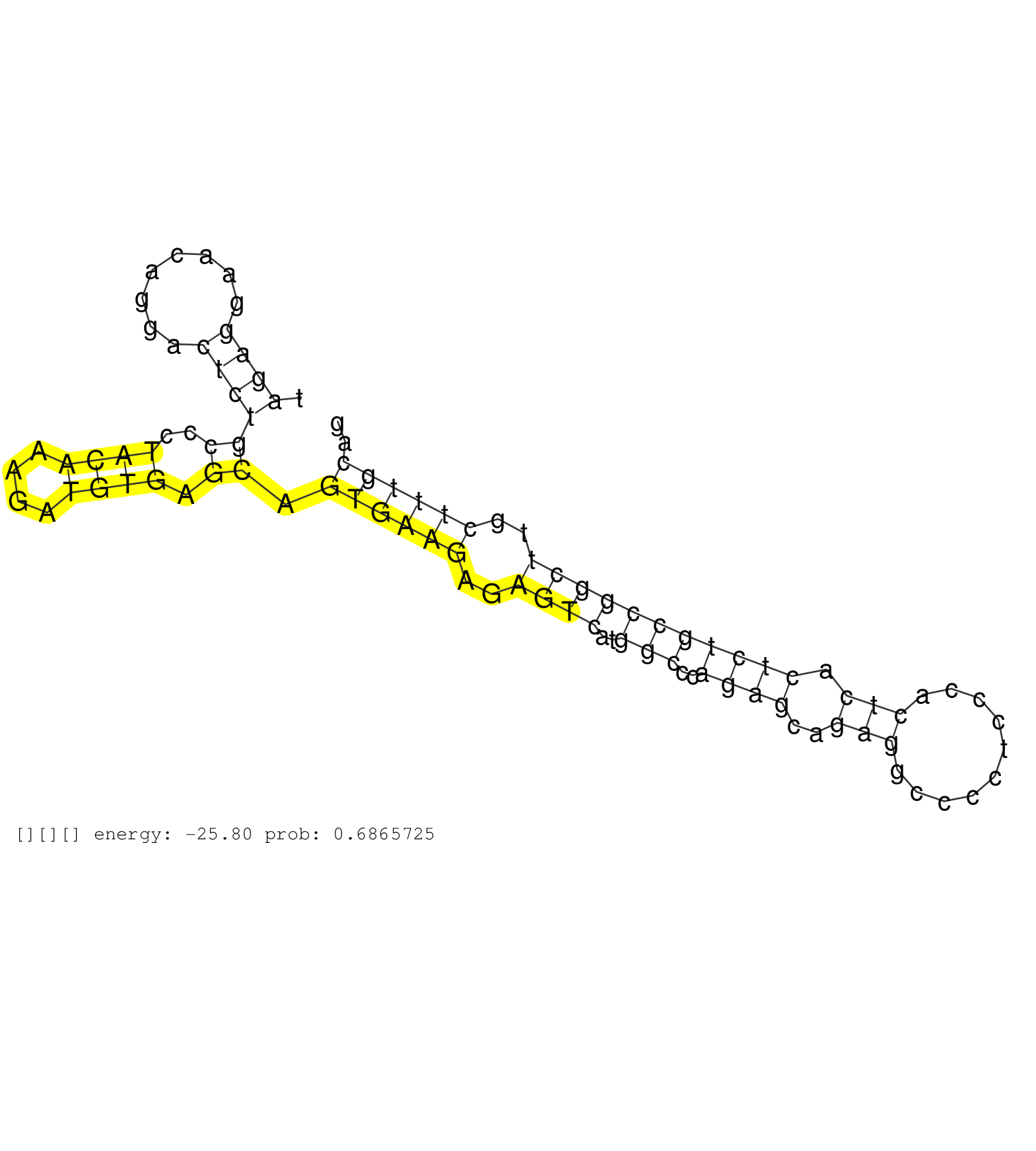
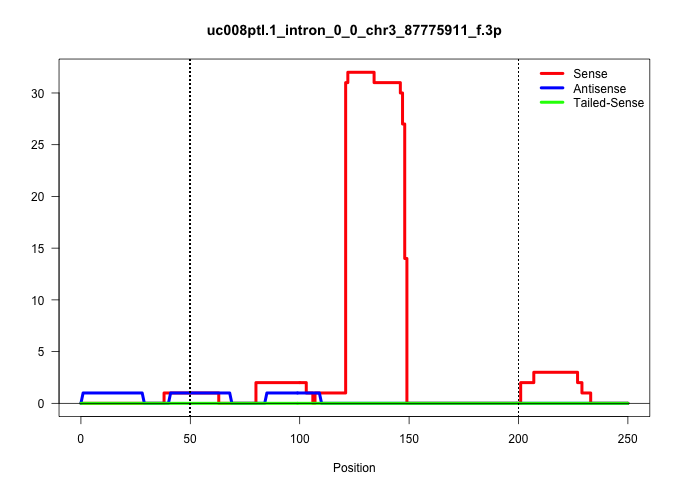
| Gene: Nes | ID: uc008ptl.1_intron_0_0_chr3_87775911_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| GCATCCTAAGAAGAAGCCTGGTCTATCTAGGAGAGGCCTGGAGTGTTTCCTGTGCTTGTCCGGGGGTCCAGCACAGTGCTCCAGACAGGGAGAACGCAGGTAGAGGAACAGGACTCTGCCCTACAAAGATGTGAGCAGTGAAGAGAGTCATGGCCCAGAGCAGAGGCCCCTCCCACTCACTCTGCCGGCTTGCTTTGCAGCTGGCTGTGGAAGCCCTGGAGCAGGAGAAGCAGGGTCTACAGAGTCAGAT .....................................................................................................((((........))))((..((((....)))).)).((((((..((((..(((..((((..(((..........))).)))))))))))..)))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................TACAAAGATGTGAGCAGTGAAGAGAGT...................................................................................................... | 27 | 1 | 13.00 | 13.00 | 2.00 | 6.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................TACAAAGATGTGAGCAGTGAAGAGAGTC..................................................................................................... | 28 | 1 | 13.00 | 13.00 | 7.00 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................GCAGCTGGCTGTGGAAGCCCTGGAGCAGG......................... | 29 | 1 | 8.00 | 8.00 | - | - | - | - | - | 8.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGCTGGCTGTGGAAGCCCTGGAG............................. | 23 | 1 | 8.00 | 8.00 | - | - | - | - | 8.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TACAAAGATGTGAGCAGTGAAGAGAG....................................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................CCAGACAGGGAGAACGCAGGTAGAGG................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGGAAGCCCTGGAGCAGGAGAAGC................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................ACAAAGATGTGAGCAGTGAAGAGAGTC..................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGGCTGTGGAAGCCCTGGAGCAGGAG....................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGCTGGCTGTGGAAGCCCTGGA.............................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................CAGACAGGGAGAACGCAGGTAGAGGA............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................ACAGGACTCTGCCCTACAAAGATGTGA.................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TACAAAGATGTGAGCAGTGAAGAGA........................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TGGAGTGTTTCCTGTGCTTGTCCGG........................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGGAAGCCCTGGAGCAGGAGAAGCAG................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGGCTGTGGAAGCCCTGGAGCAGGAGAA..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......TAAGAAGAAGCCTGGTCTATCTAGGA.......................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................CCAGACAGGGAGAACGCAGGTAG................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................................CTGTGGAAGCCCTGGAGCAGGAGAAGC................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| GCATCCTAAGAAGAAGCCTGGTCTATCTAGGAGAGGCCTGGAGTGTTTCCTGTGCTTGTCCGGGGGTCCAGCACAGTGCTCCAGACAGGGAGAACGCAGGTAGAGGAACAGGACTCTGCCCTACAAAGATGTGAGCAGTGAAGAGAGTCATGGCCCAGAGCAGAGGCCCCTCCCACTCACTCTGCCGGCTTGCTTTGCAGCTGGCTGTGGAAGCCCTGGAGCAGGAGAAGCAGGGTCTACAGAGTCAGAT .....................................................................................................((((........))))((..((((....)))).)).((((((..((((..(((..((((..(((..........))).)))))))))))..)))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................GCAGGAGAAGCAGGGTCTACAGAGTCA... | 27 | 1 | 5.00 | 5.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CAGGAGAAGCAGGGTCTACAGAGTCA... | 26 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .CATCCTAAGAAGAAGCCTGGTCTATCTA............................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................AGTGTTTCCTGTGCTTGTCCGGGGGTCC..................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................GGGAGAACGCAGGTAGAGGAcctc............................................................................................................................................... | 24 | cctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................CAGGGAGAACGCAGGTAGAGGAACA............................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |