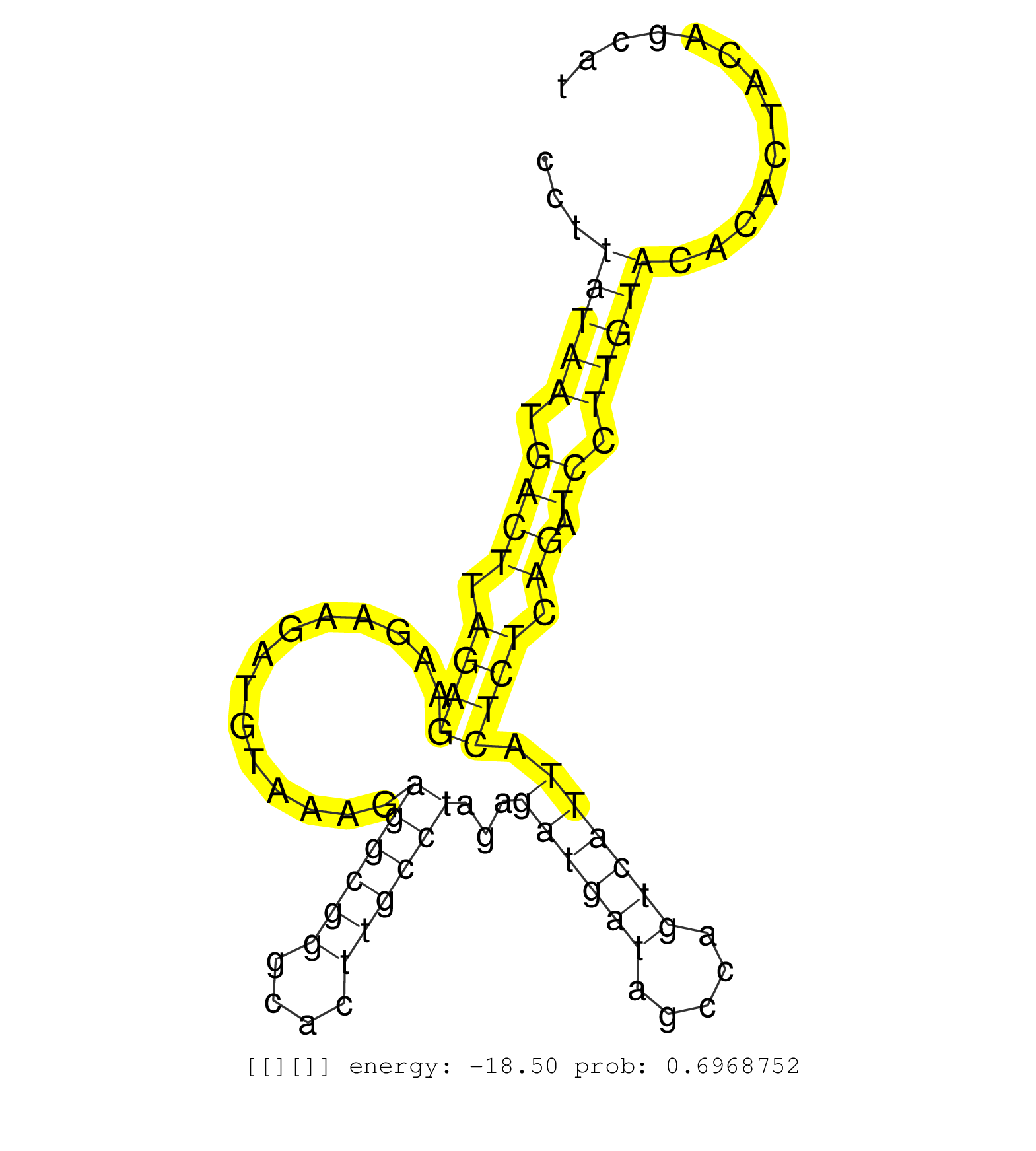

| Gene: Phf17 | ID: uc008pcj.1_intron_8_0_chr3_41408914_f.5p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(19) TESTES |
| TGTTTAGGAGGCTGCAGCTGTTCACGCACCTGCGGCAGGACCTGGAGAGGGTAATGATTGACACCGACACCTTATAATGACTTAGAGAAGAAGATGTAAAGAGGCGGGCACTTGCCTAGAGATGATAGCCAGTCATTACTCTCAGATCCTTGTACACACTACAGCATGAGGTCATGTTGGTTAAACATATTTATGGGCTGGTAAACTCACTGCATTGTGCAAGACTGCCAGTGTGGGAGTTGCTTTGTGA ........................................................................(((((.((((.((((..............((((((....))))))...((((((.....)))))).)))).)).)).)))))................................................................................................ .....................................................................70...............................................................................................167................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................TAATGACTTAGAGAAGAAGATGTAAAG..................................................................................................................................................... | 27 | 1 | 10.00 | 10.00 | 2.00 | - | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| .............................................................................TGACTTAGAGAAGAAGATGTAAAGAGGC................................................................................................................................................. | 28 | 1 | 6.00 | 6.00 | 3.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TATAATGACTTAGAGAAGAAGATGTAA....................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | - | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TATAATGACTTAGAGAAGAAGATGTAAAG..................................................................................................................................................... | 29 | 1 | 5.00 | 5.00 | - | - | 1.00 | 1.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAATGACTTAGAGAAGAAGATGTAAAGAGG.................................................................................................................................................. | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................TTTATGGGCTGGTAAACTCACTGCATTG................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................TTACTCTCAGATCCTTGTACACACTACA....................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGACTTAGAGAAGAAGATGTAAAGAGG.................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAATGACTTAGAGAAGAAGATGTAA....................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGACTTAGAGAAGAAGATGTAAAGAGGCGG............................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................ATAATGACTTAGAGAAGAAGAT........................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................AGAAGATGTAAAGAGGCGGGCACTTGC....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CACGCACCTGCGGCAGGACCTGGAG........................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................GGCAGGACCTGGAGAGGGTAATGATT............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TACTCTCAGATCCTTGTACACACTACAGCAT................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGACTTAGAGAAGAAGATGT......................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CACGCACCTGCGGCAGGACCT............................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................TAATGACTTAGAGAAGAAGATGTAAAGA.................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CAGGACCTGGAGAGGGTAATGATTG.............................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGCAAGACTGCCAGTGTGGGAGTTGC....... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TATGGGCTGGTAAACTCACTGCATT.................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................GAAGAAGATGTAAAGAGGCGGGCACTT......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................ATATTTATGGGCTGGTAAACTCACTG...................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TGATAGCCAGTCATTACTCTCAGATCC..................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTAAAGAGGCGGGCACTTGCCTAGAG................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CACTTGCCTAGAGATGATAGCCAGTa.................................................................................................................... | 26 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................TTATAATGACTTAGAGAAGAAGATGTA........................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................ATGACTTAGAGAAGAAGATGTAAAGAGG.................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CTAGAGATGATAGCCAGTCATTACTCT............................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TATTTATGGGCTGGTAAACTCACTGC..................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................TAATGACTTAGAGAAGAAGATGTAAA...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGCCAGTGTGGGAGTTGCTTT.... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TATAATGACTTAGAGAAGAAGATGTA........................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................ATGGGCTGGTAAACTCACTGCATT.................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TATAATGACTTAGAGAAGAAGATGT......................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ACTTAGAGAAGAAGATGTAAAGAGGCG................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TACTCTCAGATCCTTGTACACACTACA....................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TATGGGCTGGTAAACTCACTGCATTGT................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TTACTCTCAGATCCTTGTACACACTAC........................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAATGACTTAGAGAAGAAGATGTAAAGAG................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................AAACATATTTATGGGCTGGTAAACTCACT....................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGTTCACGCACCTGCGGCAGGACCTGG............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGACTTAGAGAAGAAGATGTAAAGAG................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAATGACTTAGAGAAGAAGATGTAAAGt.................................................................................................................................................... | 28 | t | 1.00 | 10.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .GTTTAGGAGGCTGCAGCTGTTCACG................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................CACACTACAGCATGAGGTCATGTTGGTT.................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| TGTTTAGGAGGCTGCAGCTGTTCACGCACCTGCGGCAGGACCTGGAGAGGGTAATGATTGACACCGACACCTTATAATGACTTAGAGAAGAAGATGTAAAGAGGCGGGCACTTGCCTAGAGATGATAGCCAGTCATTACTCTCAGATCCTTGTACACACTACAGCATGAGGTCATGTTGGTTAAACATATTTATGGGCTGGTAAACTCACTGCATTGTGCAAGACTGCCAGTGTGGGAGTTGCTTTGTGA ........................................................................(((((.((((.((((..............((((((....))))))...((((((.....)))))).)))).)).)).)))))................................................................................................ .....................................................................70...............................................................................................167................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|