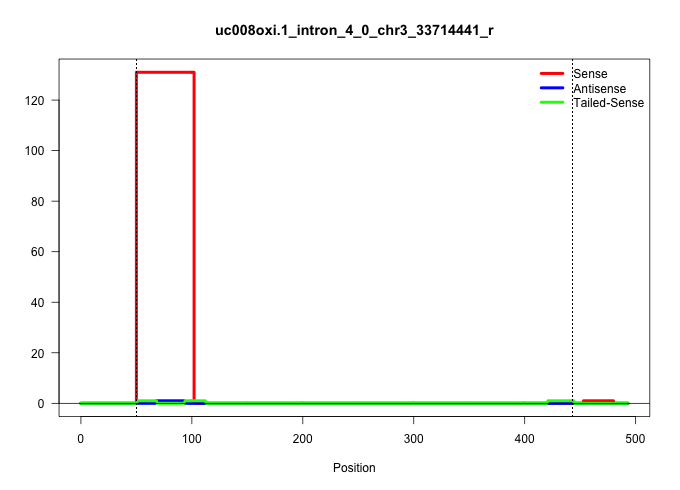
| Gene: Ccdc39 | ID: uc008oxi.1_intron_4_0_chr3_33714441_r | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(3) PIWI.ip |
(7) TESTES |
| CAGCTGCAACAGCAATTATAAGCAATCTTTCAAAAAAGTGACTCCGTCAAGTAGGTAGCAGCCATTACAACAAATATGATGACGACTAGTTAAATAAATGTAGAGAAGGAACATTATGATTCTTGGAGAATAAAAGTGTGCTATGATTGAGCCTAAAAAAAGGAATCATATTATGTTCCTCCCTCTTTGTTTTAATAAGTATGAGGTGTTATTTACATTATTAGTTTTATTTGACTATGGTATATCTCTTTAGGGAATTCGTTTTGAATATGGAAAATATACTGGTGCTATATACTAACTAAGAAATGGCTACTACATGTCTATACTGTGTTTTTCATGGGGATCTAAGATGTGTAACCTTTCATTTAAAGTCAGAGGTTTACTTGTTATTACTTTAAGGTAGCTTTGCACGTATTACATTGGAAATTAAAATTCTCTTGTAGGTGATGAGTATGCATTAAAAATTCAACTAGAAGAACAAAAAAGAACTGCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGTAGCAGCCATTACAACAAATATGATGACGACTAGTTAAATAAATGTA....................................................................................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 131.00 | 131.00 | 131.00 | - | - | - | - | - | - |
| ...................................................TAGGTAGCAGCCATgtgg........................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | gtgg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - |
| ..............................................................................................TAAATGTAGAGAAGGgtcc............................................................................................................................................................................................................................................................................................................................................................................................ | 19 | gtcc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................GGAAATTAAAATTCTCTTGTAGaa................................................ | 24 | aa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGCATTAAAAATTCAACTAGAAGAACA............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| CAGCTGCAACAGCAATTATAAGCAATCTTTCAAAAAAGTGACTCCGTCAAGTAGGTAGCAGCCATTACAACAAATATGATGACGACTAGTTAAATAAATGTAGAGAAGGAACATTATGATTCTTGGAGAATAAAAGTGTGCTATGATTGAGCCTAAAAAAAGGAATCATATTATGTTCCTCCCTCTTTGTTTTAATAAGTATGAGGTGTTATTTACATTATTAGTTTTATTTGACTATGGTATATCTCTTTAGGGAATTCGTTTTGAATATGGAAAATATACTGGTGCTATATACTAACTAAGAAATGGCTACTACATGTCTATACTGTGTTTTTCATGGGGATCTAAGATGTGTAACCTTTCATTTAAAGTCAGAGGTTTACTTGTTATTACTTTAAGGTAGCTTTGCACGTATTACATTGGAAATTAAAATTCTCTTGTAGGTGATGAGTATGCATTAAAAATTCAACTAGAAGAACAAAAAAGAACTGCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................................................................................................................................................................................TGTTATTACTTTAAgaaa............................................................................................... | 18 | gaaa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 |
| ....................................................................AACAAATATGATGACGACTAGTTAAA............................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - |