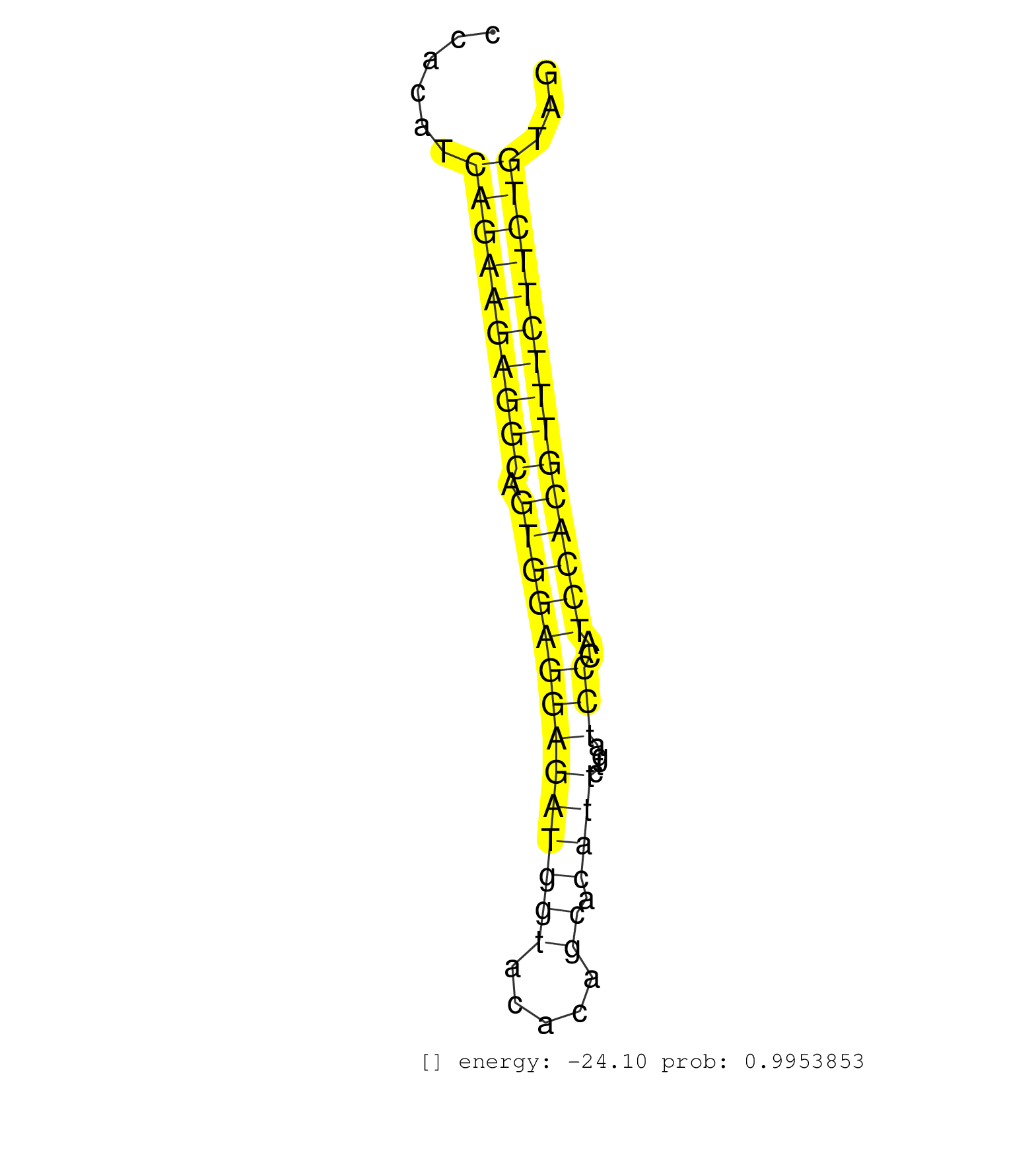

| Gene: Cpa3 | ID: uc008osp.1_intron_2_0_chr3_20122189_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.mut |
(10) TESTES |
| TTCTGTGGCCTAGAAAAGGAGGACAAATTAGGCTAAGAGTTCTCCACTAGACAGTCTGTGCACATATGTGCACAAAGCCTCTAGTGACTGTGTAACAGGAGGTACCTGGTGTTTTAAGCATCAAGTGAACCCACATCAGAAGAGGCAGTGGAGGAGATGGTACACAGCACATTCTGATCCCATCCACGTTTCTTCTGTAGCTTCTCCAAACACCAACAAACCATGCCTCAATGTCTACAGGGGACCTGCA ........................................................................................................................................((((((((((.((((((((((((((.....)).))))....)))..)))))))))))))))..................................................... ..................................................................................................................................131..................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................CCCATCCACGTTTCTTCTGTAG.................................................. | 22 | 1 | 30.00 | 30.00 | 16.00 | 6.00 | 5.00 | - | 1.00 | - | 1.00 | - | 1.00 | - |
| ..................................................................................................................................................................................CCCATCCACGTTTCTTCTGTAGt................................................. | 23 | t | 7.00 | 30.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 |
| ..................................................................................................................................................................................CCCATCCACGTTTCTTCTGTAGa................................................. | 23 | a | 3.00 | 30.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................TCAGAAGAGGCAGTGGAGGAGAT............................................................................................ | 23 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................TCAGAAGAGGCAGTGGAGGAGt............................................................................................. | 22 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TCCCATCCACGTTTCTTCTGTAG.................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ATCCCATCCACGTTgcac........................................................ | 18 | gcac | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................CCACTAGACAGTCTGgaa............................................................................................................................................................................................. | 18 | gaa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ACCATGCCTCAATGTCTACAGG......... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ATCAGAAGAGGCAGTGGAGGAG.............................................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................ATCCACGTTTCTTCTGTAG.................................................. | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CCATCCACGTTTCTTCTGTAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CCATCCACGTTTCTTCTGTA................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| TTCTGTGGCCTAGAAAAGGAGGACAAATTAGGCTAAGAGTTCTCCACTAGACAGTCTGTGCACATATGTGCACAAAGCCTCTAGTGACTGTGTAACAGGAGGTACCTGGTGTTTTAAGCATCAAGTGAACCCACATCAGAAGAGGCAGTGGAGGAGATGGTACACAGCACATTCTGATCCCATCCACGTTTCTTCTGTAGCTTCTCCAAACACCAACAAACCATGCCTCAATGTCTACAGGGGACCTGCA ........................................................................................................................................((((((((((.((((((((((((((.....)).))))....)))..)))))))))))))))..................................................... ..................................................................................................................................131..................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|