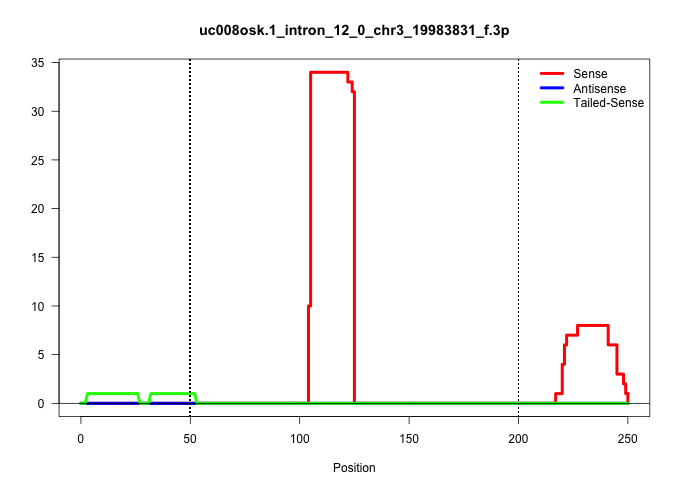
| Gene: Hltf | ID: uc008osk.1_intron_12_0_chr3_19983831_f.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| CAAAAATCTGCCTGCCTCTGCTTTCTGTGTGCTGGGATTAAAGGCAAATACCAGCATTCGCATCCCCACCTTCAGCCCATAAAATTCTTCTGGATTTCTTTCTGTGTAGATGATGATTGCTGAGACCTTATTGAGATGTATAGTTCAGGATTTTAAAAAGCAGGTCTACGAACAAGTCTGTCAATGTCTTGTTCTTTTAGGCATGTCTATGATGGAGTGCTCAGAGGCATGTGACACTGGAGAGAGGACA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................GTAGATGATGATTGCTGAGA............................................................................................................................. | 20 | 1 | 22.00 | 22.00 | 7.00 | 4.00 | 5.00 | 3.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................TGTAGATGATGATTGCTGAGA............................................................................................................................. | 21 | 1 | 10.00 | 10.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TCAGAGGCATGTGACACTGGA......... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GTAGATGATGATTGCTGAG.............................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................................................AGAGGCATGTGACACTGGAGAGAGGAC. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGCTCAGAGGCATGTGACACTGGAGAGA..... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TCAGAGGCATGTGACACTGGAGAGA..... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................GTAGATGATGATTGCTG................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CAGAGGCATGTGACACTGGAGAGA..... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................................................................CATGTGACACTGGAGAGAGGACA | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................TGGGATTAAAGGCAAATtttt..................................................................................................................................................................................................... | 21 | tttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................................................CAGAGGCATGTGACACTGGAGAGAGGA.. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...AAATCTGCCTGCCTCTGCTTTCaa............................................................................................................................................................................................................................... | 24 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| CAAAAATCTGCCTGCCTCTGCTTTCTGTGTGCTGGGATTAAAGGCAAATACCAGCATTCGCATCCCCACCTTCAGCCCATAAAATTCTTCTGGATTTCTTTCTGTGTAGATGATGATTGCTGAGACCTTATTGAGATGTATAGTTCAGGATTTTAAAAAGCAGGTCTACGAACAAGTCTGTCAATGTCTTGTTCTTTTAGGCATGTCTATGATGGAGTGCTCAGAGGCATGTGACACTGGAGAGAGGACA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAAATCTGCCTGCCTCTaaaa....................................................................................................................................................................................................................................... | 21 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |