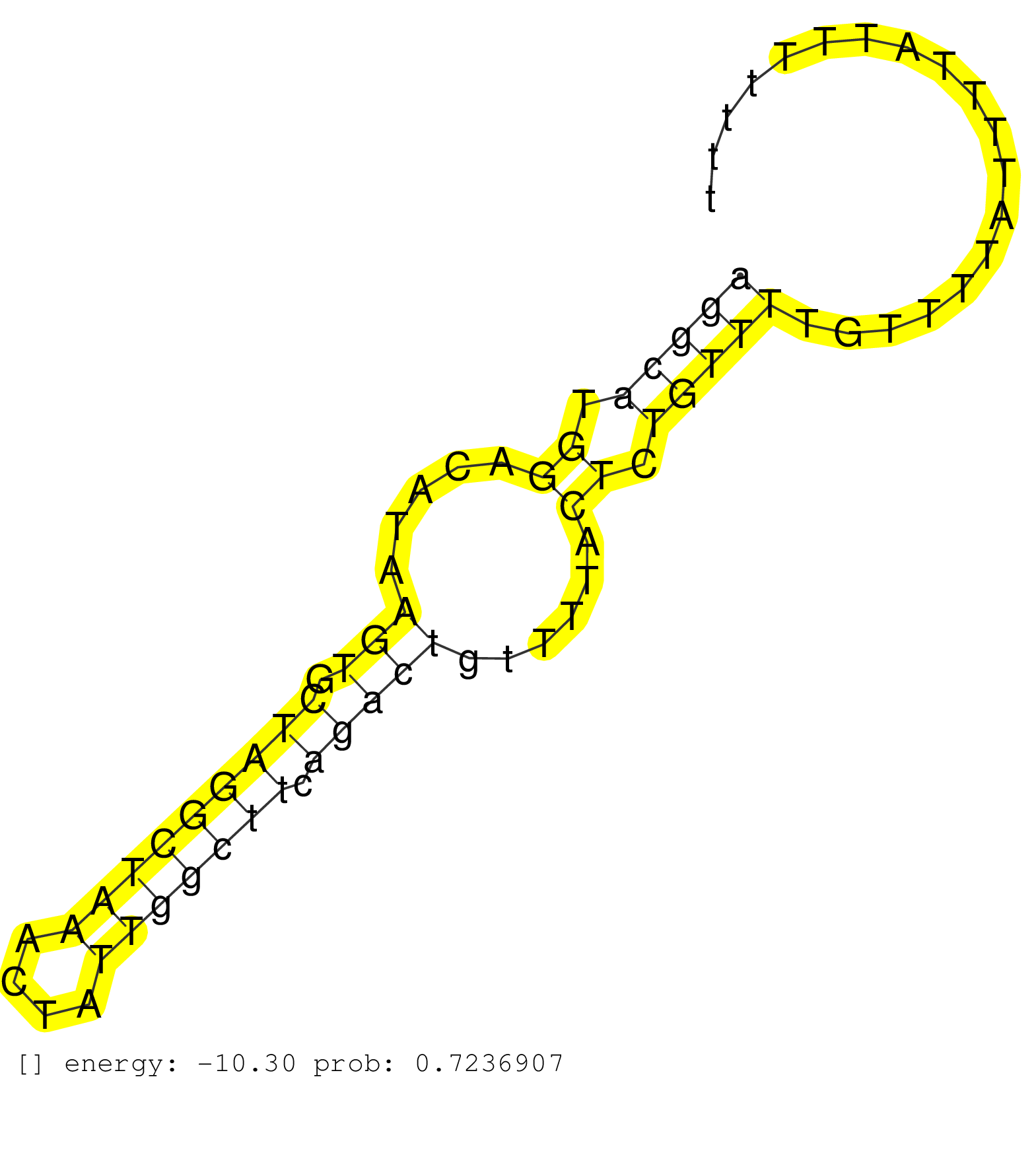
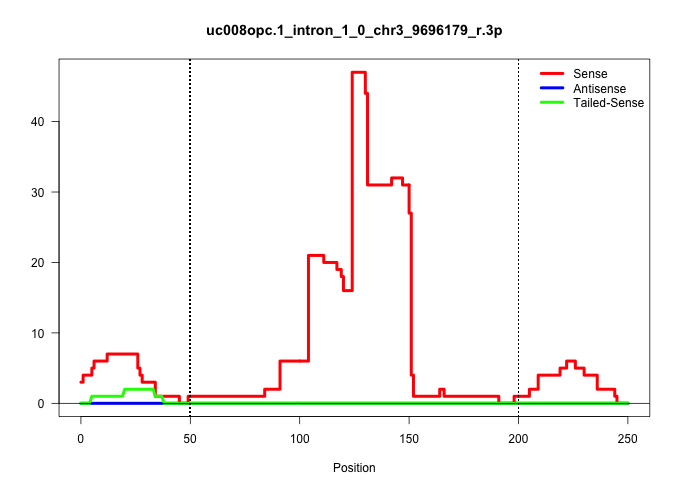
| Gene: Pag1 | ID: uc008opc.1_intron_1_0_chr3_9696179_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| AAGACCTCTGTACACCGTGAAGCCGCCAGGCAGCCATGCGCAGGCACGGTCATGGGGTGCTGATGCAGGCGACGCAGAGCTGCCTGTATCCTGTAGAAATACCGTGAAAGCATTTGCATAGGCATGGACATAAGTGCTAGGCTAAACTATTGGCTTCAGACTGTTTTACTCTGTTTTGTTTTATTTTATTTTTTTGACAGAGATTTAGTTCCTTGTCATACAAGTCTCGGGAAGAAGACCCGACTCTTAC .......................................................................................................................(((((.((.....(((.(((((((((....))))))).)))))......)).))))).......................................................................... .......................................................................................................................120........................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................TGGACATAAGTGCTAGGCTAAACTATT................................................................................................... | 27 | 1 | 23.00 | 23.00 | 6.00 | 7.00 | 3.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGAAAGCATTTGCATAGGCATGGACAT....................................................................................................................... | 27 | 1 | 12.00 | 12.00 | 5.00 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGACATAAGTGCTAGGCTAAACTAT.................................................................................................... | 26 | 1 | 4.00 | 4.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGAAAGCATTTGCATAGGCATGGACA........................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGACATAAGTGCTAGGCTAAACTATTG.................................................................................................. | 28 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGTAGAAATACCGTGAAAGCATTTGCATA.................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TCCTTGTCATACAAGTCTCGGGAAGAA.............. | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CTGATGCAGGCGACGCAGAGCTGCCTGTATCCTGTAGAA........................................................................................................................................................ | 39 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................GAAAGCATTTGCATAGGCATGGACAT....................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................AGTCTCGGGAAGAAGACCCGAC...... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ACAAGTCTCGGGAAGAAGACCCGACT..... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................TGTAGAAATACCGTGAAAGCATTTGCAT................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............CACCGTGAAGCCGCCAGGCAGCCATGCGCAGGC............................................................................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................................................................................................AGAGATTTAGTTCCTTGTCATACAAGTC........................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TAAACTATTGGCTTCAGACTGTTT.................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .AGACCTCTGTACACCGTGAAGCCGCCA.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TAGTTCCTTGTCATACAAGTCTCGG.................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGACATAAGTGCTAGGCTAAAC....................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CTCTGTACACCGTGAAGCCGCCAGGCAGt........................................................................................................................................................................................................................ | 29 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TTTACTCTGTTTTGTTTTATTTTATTT........................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................AGCCGCCAGGCAGCCAaa.................................................................................................................................................................................................................... | 18 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................TGTAGAAATACCGTGAAAGCATTTGC..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................TCATGGGGTGCTGATGCAGGCGACGC............................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGTATCCTGTAGAAATACCGTGAAAGC........................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....CTCTGTACACCGTGAAGCCGCCAGGCAGC........................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TCTGTACACCGTGAAGCCGCCAGGCAGC........................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AGAGCTGCCTGTATCCTGTAGAAATACCGT................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| AAGACCTCTGTACACCGTGAAGCCGCCAGGCAGCCATGCGCAGGCACGGTCATGGGGTGCTGATGCAGGCGACGCAGAGCTGCCTGTATCCTGTAGAAATACCGTGAAAGCATTTGCATAGGCATGGACATAAGTGCTAGGCTAAACTATTGGCTTCAGACTGTTTTACTCTGTTTTGTTTTATTTTATTTTTTTGACAGAGATTTAGTTCCTTGTCATACAAGTCTCGGGAAGAAGACCCGACTCTTAC .......................................................................................................................(((((.((.....(((.(((((((((....))))))).)))))......)).))))).......................................................................... .......................................................................................................................120........................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|