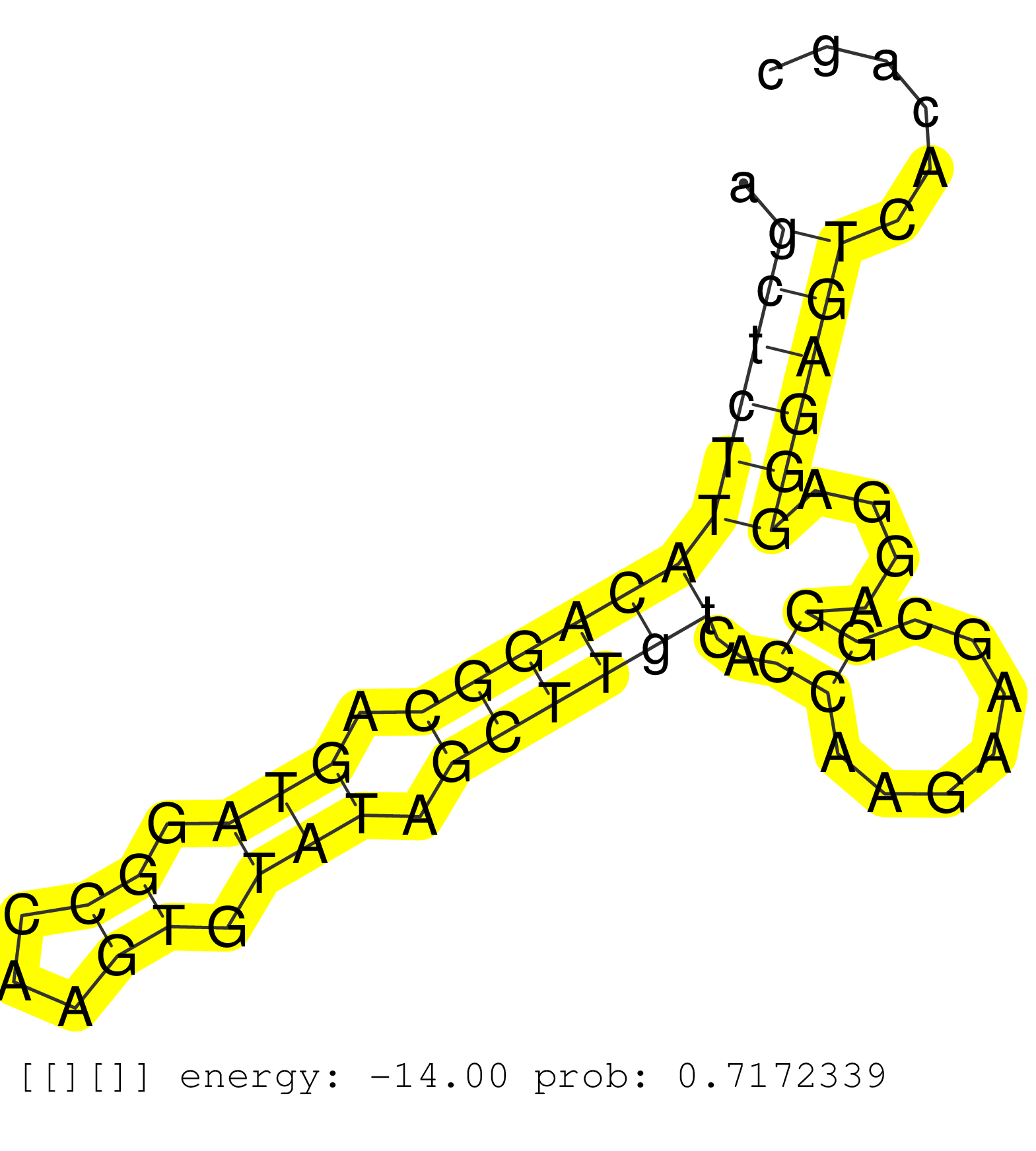
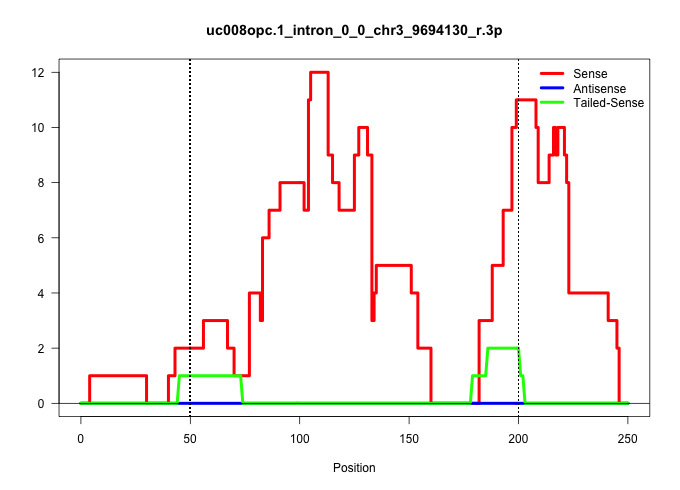
| Gene: Pag1 | ID: uc008opc.1_intron_0_0_chr3_9694130_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(13) TESTES |
| CCCATGAGATCTCCACGAAACTGGGTTTATAGAGAGTTAATCGAATAAGTCTCATCTACAGAAAGGAAGATATTGTTTAATTATTCTCTAGTAACATACAGCTCTTACAGGCAGTAGGCCAAGTGTATAGCTTGTCACCAAGAAGCGGAGGAGGGAGTCACAGCTTGCAGGTGGCTTGTTCATGTCTCTCTGTTCCCTAGATCTCAGCAATGTATTCCTCAGTGAATAAGCCTGGCCAGTCGGCACACAA ....................................................................................................((((((((((((.(((.((...)).))).))))))..((.......))....))))))............................................................................................ ...................................................................................................100..............................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................TTACAGGCAGTAGGCCAAGTGTATAGCTT..................................................................................................................... | 29 | 1 | 4.00 | 4.00 | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGATCTCAGCAATGTATTCCTCAGT........................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................TTCTCTAGTAACATACAGCTCTTACAGGCA......................................................................................................................................... | 30 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TACAGGCAGTAGGCCAAGTGTATAGCTT..................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGTCTCTCTGTTCCCTAGATCTCAGCA......................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................TAATTATTCTCTAGTAACATACAGCTCT................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CACCAAGAAGCGGAGGAGGGAGTCA.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................TAGCTTGTCACCAAGAAGCGGAGGAGG................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TACAGGCAGTAGGCCAAGTGTATAGC....................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TAACATACAGCTCTTACAGGCAGTAGG.................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TCACCAAGAAGCGGAGGAGGGAGTCA.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TCAGTGAATAAGCCTGGCCAGTCGGCAC.... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TCTCTGTTCCCTAGcgg............................................... | 17 | cgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................TCTGTTCCCTAGATCTCAGCAATGTATTC................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TCCCTAGATCTCAGCAATGTATTCCTCA............................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................AATAAGTCTCATCTACAGAAAGGAAGA.................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................TAAGTCTCATCTACAGAAAGGAAGAcatt................................................................................................................................................................................ | 29 | catt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TTCCTCAGTGAATAAGCCTGGCCAGTC......... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CCTCAGTGAATAAGCCTGGCCAGTCGGCAC.... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TATAGCTTGTCACCAAGAAGCGGAGG................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TCCCTAGATCTCAGCAATGTATTCCTCAG............................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................TACAGAAAGGAAGATATTGTTTAATT........................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................TATAGCTTGTCACCAAGAAGCGGAGGAGG................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TCTGTTCCCTAGATCTCAGCAATGTAT................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TCCTCAGTGAATAAGCCTGGCCAGTCGGCA..... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................TCGAATAAGTCTCATCTACAGAAAGGA....................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................TCTAGTAACATACAGCTCTTACAGGCAGT....................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................TAATTATTCTCTAGTAACATACAGC.................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATCTCAGCAATGTATTCCTCAGT........................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....TGAGATCTCCACGAAACTGGGTTTAT............................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................TGTCTCTCTGTTCCCTAGATCTCAGC.......................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................TCATGTCTCTCTGTTCCCTAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| CCCATGAGATCTCCACGAAACTGGGTTTATAGAGAGTTAATCGAATAAGTCTCATCTACAGAAAGGAAGATATTGTTTAATTATTCTCTAGTAACATACAGCTCTTACAGGCAGTAGGCCAAGTGTATAGCTTGTCACCAAGAAGCGGAGGAGGGAGTCACAGCTTGCAGGTGGCTTGTTCATGTCTCTCTGTTCCCTAGATCTCAGCAATGTATTCCTCAGTGAATAAGCCTGGCCAGTCGGCACACAA ....................................................................................................((((((((((((.(((.((...)).))).))))))..((.......))....))))))............................................................................................ ...................................................................................................100..............................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................TAATCGAATAAGTgatg........................................................................................................................................................................................................ | 17 | gatg | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |