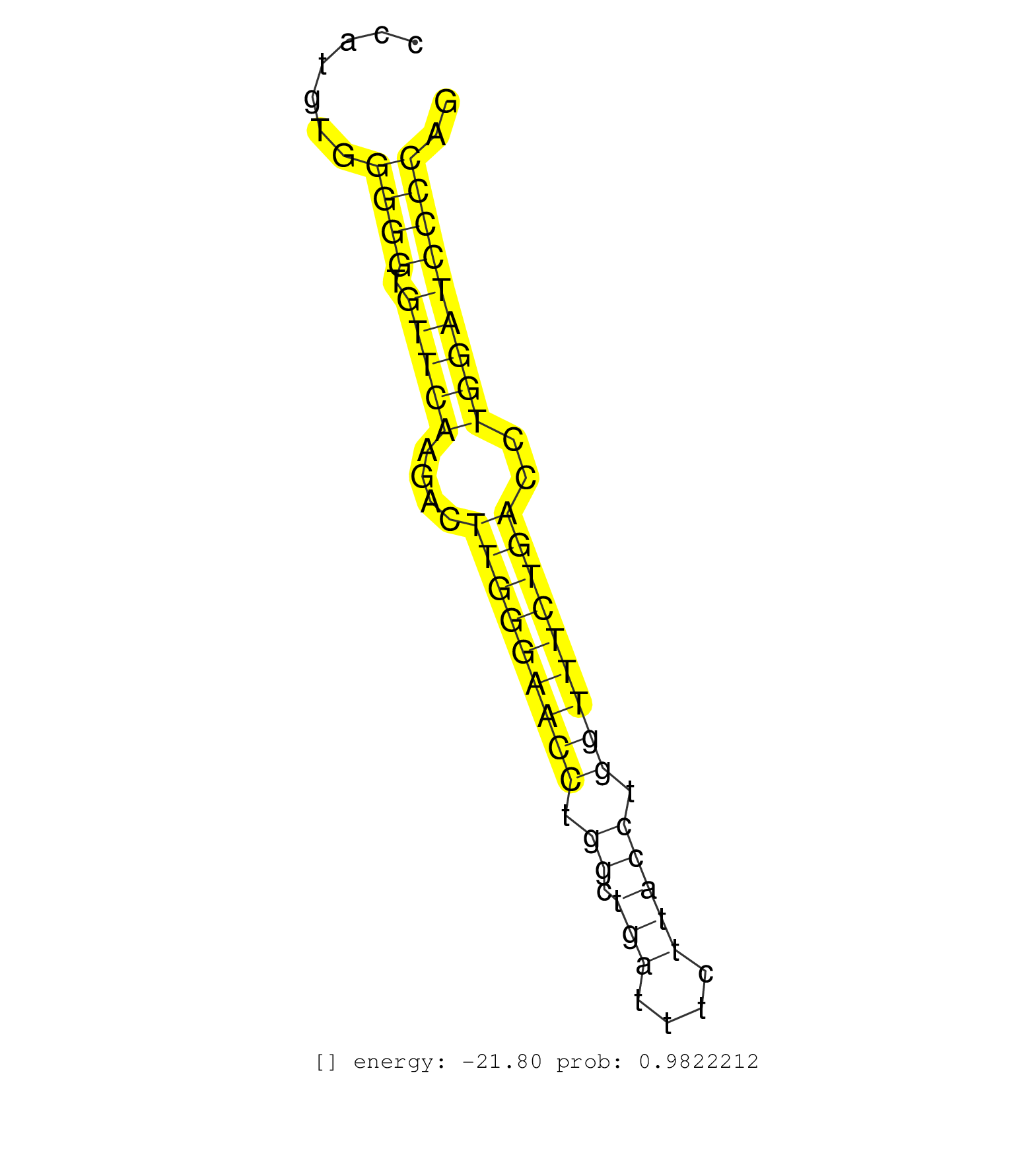
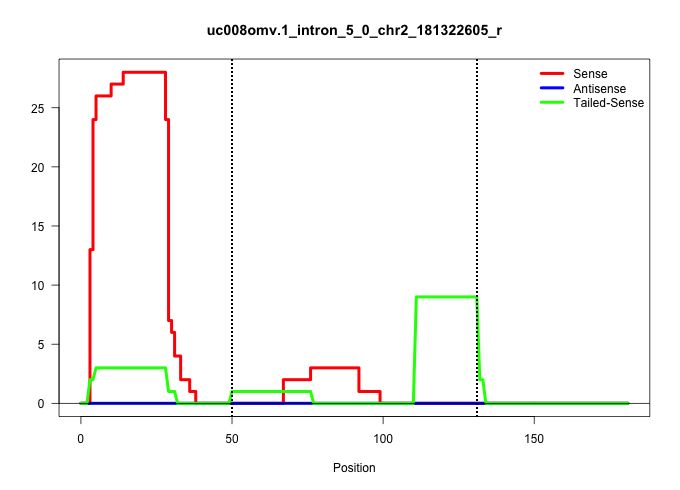
| Gene: Gm632 | ID: uc008omv.1_intron_5_0_chr2_181322605_r | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| GGCTTCGAAAGGTGCTGAAGCAGATGGGAAGGCTGCGCTGCCCCCAAGAGGTCAGTGTTGGCCCATGTGGGGGTGTTCAAGACTTGGGAACCTGGCTGATTTCTTACCTGGTTTCTGACCTGGATCCCCAGGGCTGTGGGGCTGCCTTCTCCAGCCTCATGGGTTATCAATACCACCAGCG .....................................................................((((.(((((....(((((((((.((.(((....))))).)))))))))..))))))))).................................................... ..............................................................63..................................................................131................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...TTCGAAAGGTGCTGAAGCAGATGGGA........................................................................................................................................................ | 26 | 1 | 9.00 | 9.00 | 1.00 | - | 2.00 | 2.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TTTCTGACCTGGATCCCCAGt................................................. | 21 | t | 8.00 | 0.00 | - | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - |
| ....TCGAAAGGTGCTGAAGCAGATGGGA........................................................................................................................................................ | 25 | 1 | 8.00 | 8.00 | 4.00 | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...TTCGAAAGGTGCTGAAGCAGATGGG......................................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| ...................................................................TGGGGGTGTTCAAGACTTGGGAACC......................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ...............................................................................................................TTTCTGACCTGGATCCCCAGaat............................................... | 23 | aat | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CAAGACTTGGGAACCTGGCTGA.................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TTCGAAAGGTGCTGAAGCAGATGGGt........................................................................................................................................................ | 26 | t | 2.00 | 4.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TTTCTGACCTGGATCCCCAGa................................................. | 21 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........GGTGCTGAAGCAGATGGGAAGGCTGCGC............................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTCAGTGTTGGCCCATGTGGGGGggtt........................................................................................................ | 27 | ggtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....TCGAAAGGTGCTGAAGCAGATGGGAA....................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCGAAAGGTGCTGAAGCAGATGGGAAG...................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CGAAAGGTGCTGAAGCAGATGGGAAGt..................................................................................................................................................... | 27 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....CGAAAGGTGCTGAAGCAGATGGGAAGGC.................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TCAAGACTTGGGAACCTGGCTGA.................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....TCGAAAGGTGCTGAAGCAGATGGGAAGGC.................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CTGAAGCAGATGGGAAGGCTGC................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....CGAAAGGTGCTGAAGCAGATGGGAAG...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGCTTCGAAAGGTGCTGAAGCAGATGGGAAGGCTGCGCTGCCCCCAAGAGGTCAGTGTTGGCCCATGTGGGGGTGTTCAAGACTTGGGAACCTGGCTGATTTCTTACCTGGTTTCTGACCTGGATCCCCAGGGCTGTGGGGCTGCCTTCTCCAGCCTCATGGGTTATCAATACCACCAGCG .....................................................................((((.(((((....(((((((((.((.(((....))))).)))))))))..))))))))).................................................... ..............................................................63..................................................................131................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|