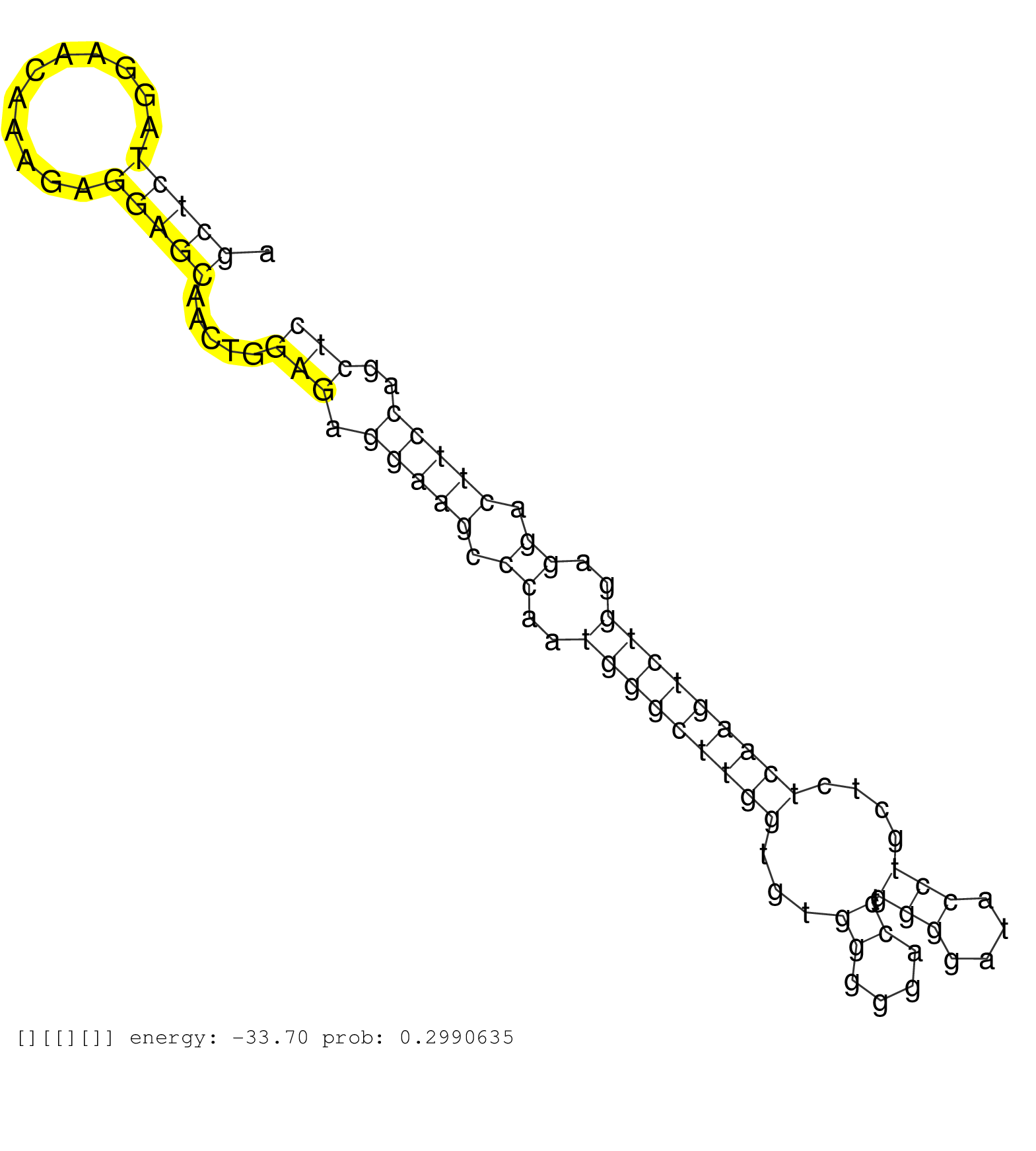
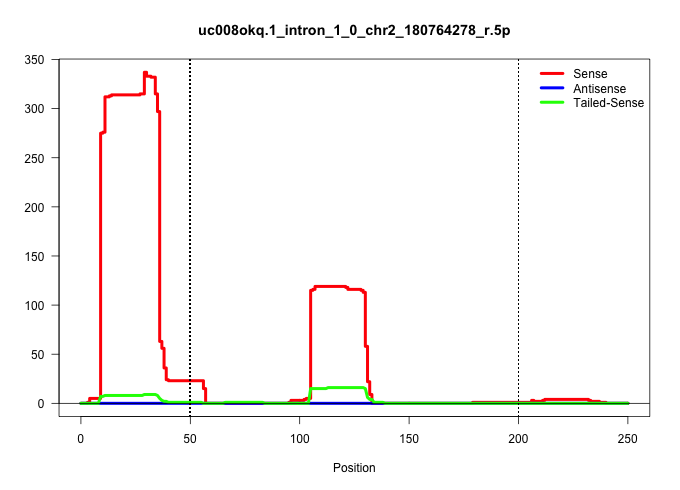
| Gene: Chrna4 | ID: uc008okq.1_intron_1_0_chr2_180764278_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(17) TESTES |
| CGCATCCCATCTGAACTCATCTGGAGGCCTGACATCGTCCTCTACAACAAGTAAGCCGTGTGGGCCCTGGCAGGTGAGATCAGGCCCCACCCAGCAAGGAAGCTCTAGGAACAAAGAGGAGCAACTGGAGAGGAAGCCCAATGGGCTTGGTGTGGGGGACCTGGGGATACCTGCTCTCAAGTCTGGAGGACTTCCAGCTCTGCTTGTCTCCTAAGGGCAGAGAATGGATTGGACATCCTCCTCTGGCAGT .....................................................................................................(((((...........))))).....(((.(((((.((..(((((((((...((....)).(((....)))....)))))))))..)).)))))..))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........TCTGAACTCATCTGGAGGCCTGACATC...................................................................................................................................................................................................................... | 27 | 1 | 231.00 | 231.00 | 74.00 | 83.00 | 32.00 | 33.00 | - | - | 6.00 | 3.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGAG........................................................................................................................ | 25 | 1 | 54.00 | 54.00 | 16.00 | 17.00 | 6.00 | - | 6.00 | 6.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGAGA....................................................................................................................... | 26 | 1 | 36.00 | 36.00 | 7.00 | 11.00 | 1.00 | 4.00 | 8.00 | 2.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - |
| ...........TGAACTCATCTGGAGGCCTGACATCGT.................................................................................................................................................................................................................... | 27 | 1 | 18.00 | 18.00 | 9.00 | 4.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACAT....................................................................................................................................................................................................................... | 26 | 1 | 17.00 | 17.00 | 11.00 | 4.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACA........................................................................................................................................................................................................................ | 25 | 1 | 15.00 | 15.00 | 2.00 | 6.00 | 2.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| .............................TGACATCGTCCTCTACAACAAGTAAGCC................................................................................................................................................................................................. | 28 | 1 | 15.00 | 15.00 | 9.00 | 1.00 | - | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGAGAG...................................................................................................................... | 27 | 1 | 11.00 | 11.00 | 2.00 | 2.00 | - | 1.00 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGAGt....................................................................................................................... | 26 | t | 8.00 | 54.00 | 2.00 | 1.00 | - | - | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................TGACATCGTCCTCTACAACAAGTAAGC.................................................................................................................................................................................................. | 27 | 1 | 7.00 | 7.00 | 4.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGAACTCATCTGGAGGCCTGACATCGTC................................................................................................................................................................................................................... | 28 | 1 | 7.00 | 7.00 | 3.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGAGAGG..................................................................................................................... | 28 | 1 | 7.00 | 7.00 | 1.00 | - | - | - | 2.00 | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........TGAACTCATCTGGAGGCCTGACATCG..................................................................................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | 2.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACATCGTC................................................................................................................................................................................................................... | 30 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........TGAACTCATCTGGAGGCCTGACATC...................................................................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| ....TCCCATCTGAACTCATCTGGAGGCCT............................................................................................................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGAGAGGt.................................................................................................................... | 29 | t | 3.00 | 7.00 | - | - | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................GGAACAAAGAGGAGCAACTGGAGAGG..................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACATCGT.................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGAACTCATCTGGAGGCCTGACA........................................................................................................................................................................................................................ | 23 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................AGGAAGCTCTAGGAACAAAGAGGAGC................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACATa...................................................................................................................................................................................................................... | 27 | a | 2.00 | 17.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................AGTCTGGAGGACTTCCAGCTCTGCTTGT........................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................AAGAGGAGCAACTGGAGAGGAAGCgc............................................................................................................... | 26 | gc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................AGGAACAAAGAGGAGCAACTGGAGAG...................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............AACTCATCTGGAGGCCTGACATCGTCC.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TCTAGGAACAAAGAGGAGCAACTGGA......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................CTGGCAGGTGAGATacca...................................................................................................................................................................... | 18 | acca | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............ACTCATCTGGAGGCCTGACATCGTC................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCCCATCTGAACTCATCTGGAGGCCTGA.......................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGAaa....................................................................................................................... | 26 | aa | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................GGAACAAAGAGGAGCAACTGGAGAG...................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTcgag........................................................................................................................ | 25 | cgag | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................CCTGACATCGTCCTCTACAACAAGTAAGC.................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACATCa..................................................................................................................................................................................................................... | 28 | a | 1.00 | 231.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACATCG..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................CTCTAGGAACAAAGAGGAGCAACTGGAG........................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACATCaa.................................................................................................................................................................................................................... | 29 | aa | 1.00 | 231.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...ATCCCATCTGAACTCATCTGGAGGCCT............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGAACTCATCTGGAGGCCTGACAT....................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................................................TAAGGGCAGAGAATGGATTGGACATCCTC.......... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................AAGGAAGCTCTAGGAACAAAGAGGAG................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TGACATCGTCCTCTACAACAAGTgaag.................................................................................................................................................................................................. | 27 | gaag | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGAGAa...................................................................................................................... | 27 | a | 1.00 | 36.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGG.......................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACATCt..................................................................................................................................................................................................................... | 28 | t | 1.00 | 231.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGAACTCATCTGGAGGCCTGACATCGTCa.................................................................................................................................................................................................................. | 29 | a | 1.00 | 7.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGA......................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................AAGGGCAGAGAATGGATTGGACATC............. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........CTGAACTCATCTGGAGGCCTGACATCG..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGACATt...................................................................................................................................................................................................................... | 27 | t | 1.00 | 17.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCTCCTAAGGGCAGAGAATGGATTGGA................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCTCCTAAGGGCAGAGAATGGATTGG.................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........TCTGAACTCATCTGGAGGCCTGAtat....................................................................................................................................................................................................................... | 26 | tat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGAACAAAGAGGAGCAACTGGAGta...................................................................................................................... | 27 | ta | 1.00 | 54.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CGCATCCCATCTGAACTCATCTGGAGGCCTGACATCGTCCTCTACAACAAGTAAGCCGTGTGGGCCCTGGCAGGTGAGATCAGGCCCCACCCAGCAAGGAAGCTCTAGGAACAAAGAGGAGCAACTGGAGAGGAAGCCCAATGGGCTTGGTGTGGGGGACCTGGGGATACCTGCTCTCAAGTCTGGAGGACTTCCAGCTCTGCTTGTCTCCTAAGGGCAGAGAATGGATTGGACATCCTCCTCTGGCAGT .....................................................................................................(((((...........))))).....(((.(((((.((..(((((((((...((....)).(((....)))....)))))))))..)).)))))..))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|