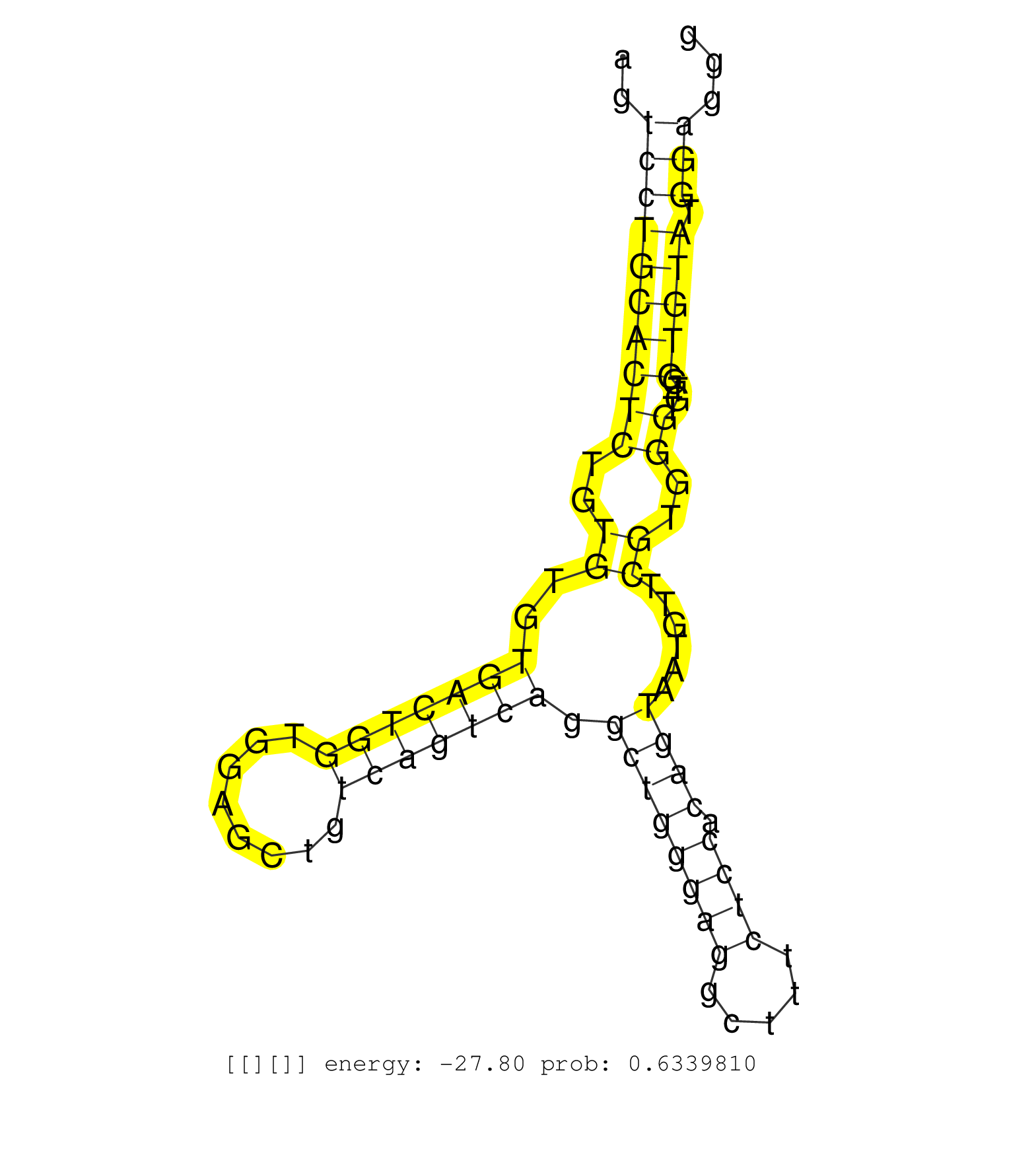

| Gene: Chrna4 | ID: uc008okq.1_intron_1_0_chr2_180764278_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(14) TESTES |
| CACCTTGACTCAAGGGTGAGAAACTGAAGTGCACAGGTGAGTGACGGCCCAGTCCACATGGACCACCACACACAGTCCTGCACTCTGTGTGTGACTGGTGGAGCTGTCAGTCAGGCTGGGAGGCTTTCTCCACAGTAATGTTCGTGGGTGTGGTGTATGGAGGGGCAGACCTCATTGCTCTTTCTCTCGCTTCCTGCCAGCGCGGACGGGGACTTTGCAGTCACCCACCTAACCAAAGCCCACCTGTTCT ...........................................................................((((((((((..((..(((((((........))))))).((((((((.....)))).))))......))..))....))))).)))......................................................................................... .........................................................................74........................................................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................TGAGAAACTGAAGTGCACAGGTGAGT................................................................................................................................................................................................................ | 26 | 1 | 33.00 | 33.00 | 12.00 | 7.00 | 5.00 | 2.00 | 2.00 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - |
| ..............................................................................TGCACTCTGTGTGTGACTGGTGGAGC.................................................................................................................................................. | 26 | 1 | 19.00 | 19.00 | 8.00 | 9.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| .....TGACTCAAGGGTGAGAAACTGAAGTGC.......................................................................................................................................................................................................................... | 27 | 1 | 8.00 | 8.00 | 4.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .....TGACTCAAGGGTGAGAAACTGAAGTG........................................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - |
| .................GAGAAACTGAAGTGCACAGGTGAGT................................................................................................................................................................................................................ | 25 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................TAATGTTCGTGGGTGTGGTGTATGG.......................................................................................... | 25 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ................TGAGAAACTGAAGTGCACAGGTGAGTGACGGC.......................................................................................................................................................................................................... | 32 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........CAAGGGTGAGAAACTGAAGTGCACAGGT.................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TGCCAGCGCGGACGGGGACTTTGCAG.............................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................TGCCAGCGCGGACGGGGACTTTGC................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........CAAGGGTGAGAAACTGAAGTGCAC........................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................CACTCTGTGTGTGACTGGTGGAGC.................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........CAAGGGTGAGAAACTGAAGTGCACAGG..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................TGAGAAACTGAAGTGCACAGGTGAGc................................................................................................................................................................................................................ | 26 | c | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TGAGAAACTGAAGTGCACAGGTGAGTG............................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGACTCAAGGGTGAGAAACTGAAGTGCAC........................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCGTGGGTGTGGTGTATGGAGGGGCAt.................................................................................. | 27 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................AATGTTCGTGGGTGTGGTGTATGGA......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................TGCACTCTGTGTGTGACTGGTGGA.................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TGAGAAACTGAAGTGCACAGGTGAG................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GAGAAACTGAAGTGCACAGGTGAGTGAC............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................AGAAACTGAAGTGCACAGGTGAGTGACGGC.......................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TCCACAGTAATGTTCGTGGGTGTGGTG............................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................CCTGCACTCTGTGTGTGACTGGTGG..................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TCCTGCACTCTGTGTGTGACTGGTGG..................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................AGAAACTGAAGTGCACAGGTGAGTGACaggc......................................................................................................................................................................................................... | 31 | aggc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACCTTGACTCAAGGGTGAGAAACTGAAGTGCACAGGTGAGTGACGGCCCAGTCCACATGGACCACCACACACAGTCCTGCACTCTGTGTGTGACTGGTGGAGCTGTCAGTCAGGCTGGGAGGCTTTCTCCACAGTAATGTTCGTGGGTGTGGTGTATGGAGGGGCAGACCTCATTGCTCTTTCTCTCGCTTCCTGCCAGCGCGGACGGGGACTTTGCAGTCACCCACCTAACCAAAGCCCACCTGTTCT ...........................................................................((((((((((..((..(((((((........))))))).((((((((.....)))).))))......))..))....))))).)))......................................................................................... .........................................................................74........................................................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................AGTCCACATGGACCAcgat......................................................................................................................................................................................... | 19 | cgat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |