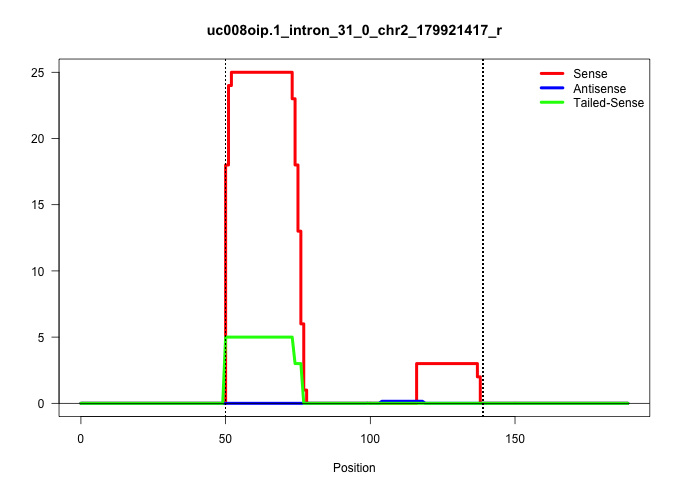
| Gene: Lama5 | ID: uc008oip.1_intron_31_0_chr2_179921417_r | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| CCTCGAGTGCGCCCCTGGCTACTGGGGCCTCCCAGAGAAGGGCTGCAGGCGTGAGTACAGGACTGTGGGAAGGCAGGGTGGAGAGAGGCAGGCATCCCTGAGCGCCTCCAGTCACATCTGCCTACCCCTCTTCCTGCAGGCTGCCAGTGTCCCCGAGGCCACTGTGACCCACACACGGGCCACTGCACC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTACAGGACTGTGGGAAGGCA.................................................................................................................. | 25 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 |
| ..................................................GTGAGTACAGGACTGTGGGAAGGC................................................................................................................... | 24 | 1 | 5.00 | 5.00 | 1.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACAGGACTGTGGGAAGGCAG................................................................................................................. | 26 | 1 | 4.00 | 4.00 | 1.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTACAGGACTGTGGGAAGGCAGG................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - |
| ....................................................................................................................TCTGCCTACCCCTCTTCCTGCA................................................... | 22 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACAGGACTGTGGGAAGG.................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACAGGACTGTGGGAAGGCAGG................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTACAGGACTGTGGGAAGGCAG................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................TCTGCCTACCCCTCTTCCTGC.................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTACAGGACTGTGGGAAGGCAGGG............................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACAGGACTGTGGGAAGGCAta................................................................................................................ | 27 | ta | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................GAGTACAGGACTGTGGGAAGGCAG................................................................................................................. | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACAGGACTGTGGGAAGGCAGt................................................................................................................ | 27 | t | 1.00 | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGTACAGGACTGTGGGAAGGt................................................................................................................... | 24 | t | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACAGGACTGTGGGAAGGCAaa................................................................................................................ | 27 | aa | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACAGGACTGTGGGAAGcc................................................................................................................... | 24 | cc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCTCGAGTGCGCCCCTGGCTACTGGGGCCTCCCAGAGAAGGGCTGCAGGCGTGAGTACAGGACTGTGGGAAGGCAGGGTGGAGAGAGGCAGGCATCCCTGAGCGCCTCCAGTCACATCTGCCTACCCCTCTTCCTGCAGGCTGCCAGTGTCCCCGAGGCCACTGTGACCCACACACGGGCCACTGCACC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................CCTCCAGTCACATCT...................................................................... | 15 | 13 | 0.15 | 0.15 | - | 0.15 | - | - | - | - | - | - | - | - | - | - | - | - |