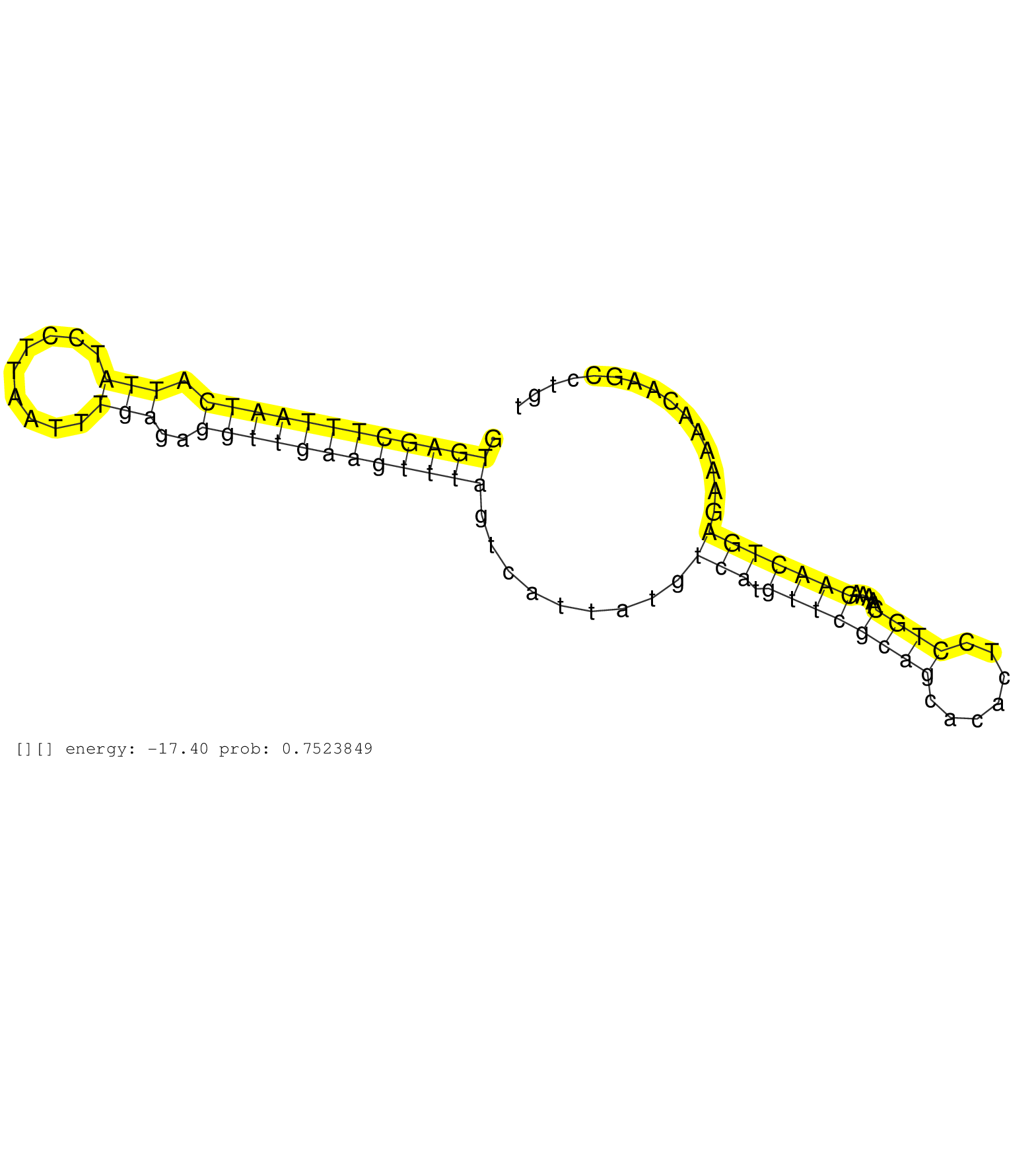
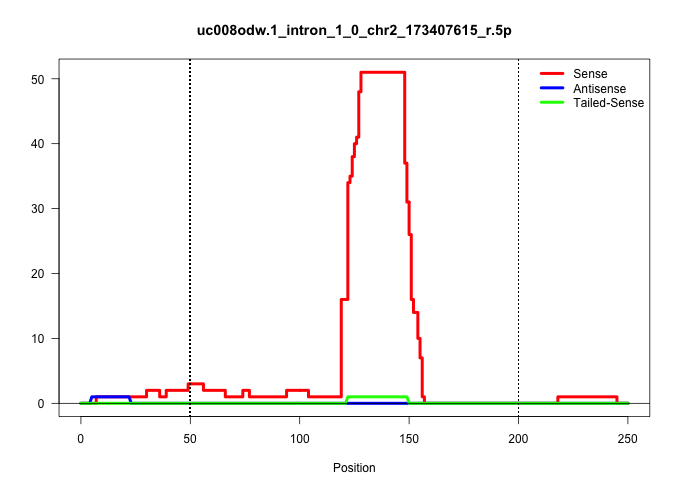
| Gene: AK029171 | ID: uc008odw.1_intron_1_0_chr2_173407615_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| GATATGATTTGGCCGGTGCCGGGGGCAAACTGGAAGATTTTCTCTTTGCTGTGAGCTTTAATCATTATCCTTAATTTGAGAGGTTGAAGTTTAGTCATTATGTCATGTTCGCAGCACACTCCTGCAAAAAGAACTGAGAAAAACAAGCCTGTGCCCGGCGCGGCGACCCTGCGCCCCCTGGCGGCGGGTGCTGGGCGAGCCCCCGGCGTCCCGGGCCCTATCTCCCATCGCTTGGCTTATTGCAGTGGCC ...................................................((((((((((((.(((.........)))..)))))))))))).........(((.((((((((.......)))).....)))))))................................................................................................................. ..................................................51...................................................................................................152................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................TCCTGCAAAAAGAACTGAGAAAAACAAGC...................................................................................................... | 29 | 1 | 12.00 | 12.00 | 5.00 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TGCAAAAAGAACTGAGAAAAACAAGCCTG................................................................................................... | 29 | 1 | 10.00 | 10.00 | 3.00 | 1.00 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................TGCAAAAAGAACTGAGAAAAACAAGCCT.................................................................................................... | 28 | 1 | 5.00 | 5.00 | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................AAAGAACTGAGAAAAACAAGCCTGTGCCC.............................................................................................. | 29 | 1 | 4.00 | 4.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TCCTGCAAAAAGAACTGAGAAAAACAAGCC..................................................................................................... | 30 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TGCAAAAAGAACTGAGAAAAACAAGCC..................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AAGAACTGAGAAAAACAAGCCTGTGCCC.............................................................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................AAAGAACTGAGAAAAACAAGCCTGTGCC............................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................AAAAGAACTGAGAAAAACAAGCCTGTGCC............................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TGCAAAAAGAACTGAGAAAAACAAGC...................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................TCATTATGTCATGTTCGCAGCACACTCC................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TGCAAAAAGAACTGAGAAAAACAAGCaa.................................................................................................... | 28 | aa | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TTCTCTTTGCTGTGAGCTTTAATCATT........................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................AAAGAACTGAGAAAAACAAGCCTGTGC................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......TTTGGCCGGTGCCGGGGGCAAACTGGAAG...................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................CAAAAAGAACTGAGAAAAACAAGCCTGT.................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AAAAAGAACTGAGAAAAACAAGCCTGT.................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AAAAAGAACTGAGAAAAACAAGCCTGTGC................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CAAAAAGAACTGAGAAAAACAAGC...................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................TTTGAGAGGTTGAAGTTTAGTCATTATGTC.................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AAGAACTGAGAAAAACAAGCCTGTGCCCG............................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................TGTGAGCTTTAATCATTATCCTTAATTT............................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CAAAAAGAACTGAGAAAAACAAGCCTGTGC................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TATCTCCCATCGCTTGGCTTATTGCAG..... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................TGGAAGATTTTCTCTTTGCTGTGAGC.................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................GCAAAAAGAACTGAGAAAAACAAGCCTGTGC................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| GATATGATTTGGCCGGTGCCGGGGGCAAACTGGAAGATTTTCTCTTTGCTGTGAGCTTTAATCATTATCCTTAATTTGAGAGGTTGAAGTTTAGTCATTATGTCATGTTCGCAGCACACTCCTGCAAAAAGAACTGAGAAAAACAAGCCTGTGCCCGGCGCGGCGACCCTGCGCCCCCTGGCGGCGGGTGCTGGGCGAGCCCCCGGCGTCCCGGGCCCTATCTCCCATCGCTTGGCTTATTGCAGTGGCC ...................................................((((((((((((.(((.........)))..)))))))))))).........(((.((((((((.......)))).....)))))))................................................................................................................. ..................................................51...................................................................................................152................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....GATTTGGCCGGTGCCGGG................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........GCCGGTGCCGGGGaca.................................................................................................................................................................................................................................. | 16 | aca | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |