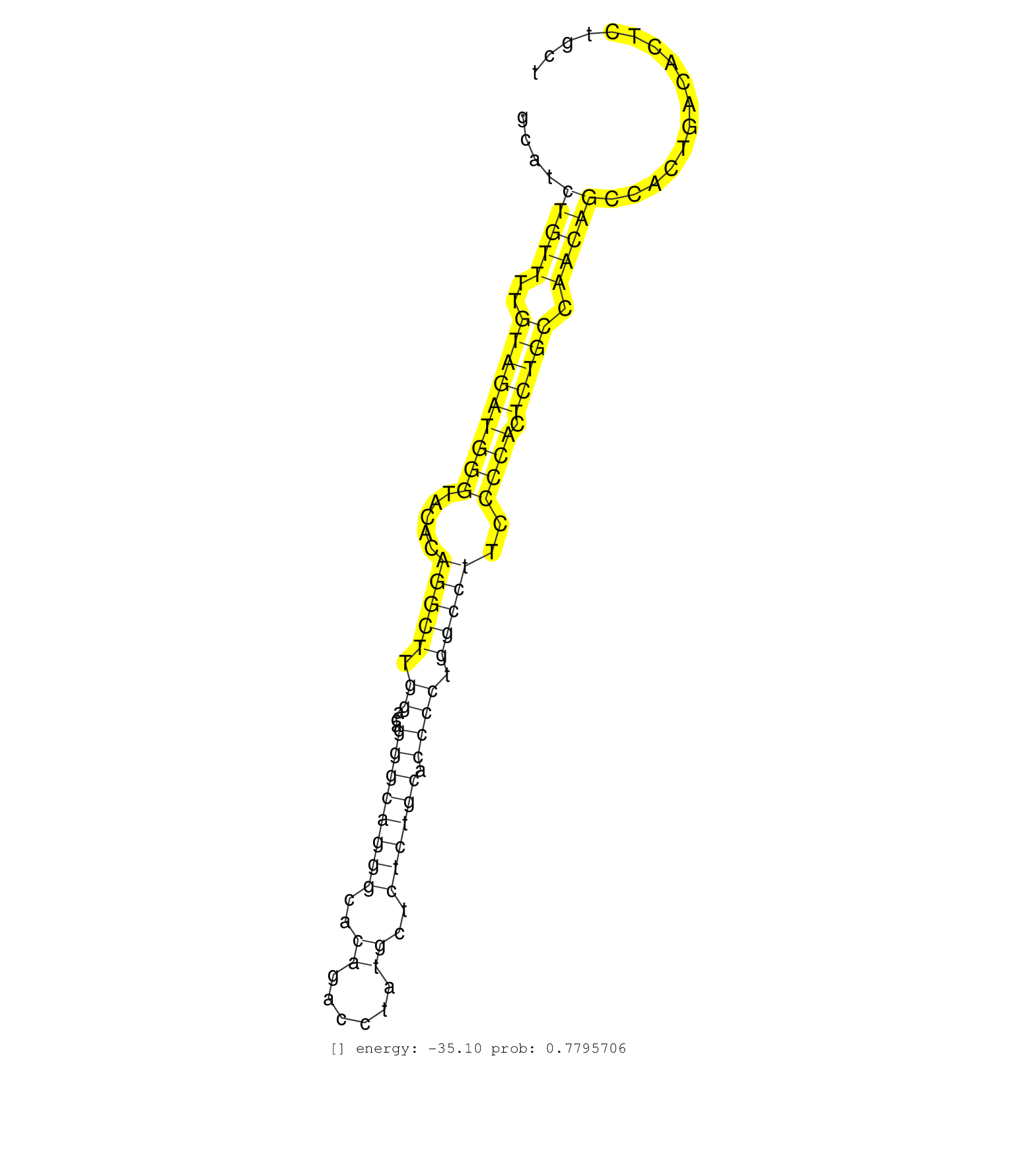

| Gene: Rbm38 | ID: uc008odh.1_intron_2_0_chr2_172848883_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| GAGCTGGCGCTAAGTGGCCTGTGGAGTGATGGCACTGTCTGTGCAAAGGCCCTGGGGTTGGTGCCCACCTGAGGTGTTGCTGCATCTGTTTTGTAGATGGGTACACAGGCTTGGACAGGGCAGGGCACAGACCTATGCTCTCTGCACCCCTGGCCTTCCCCACTCTGCCAACAGCCACTGACACTCTGCTTGGCCCACAGGCTGACTCCTCACTACATCTACCCACCAGCCATTGTGCAGCCCAGCGTGG .....................................................................................(((((..(((((((((.....(((((.((...((((((((..((......))..)))))).)))).)))))..)))).))))).)))))............................................................................ .................................................................................82..........................................................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................TGTTTTGTAGATGGGTACACAGGCTT.......................................................................................................................................... | 26 | 1 | 7.00 | 7.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| ...........................................................................................TGTAGATGGGTACACAGGCTTGGACAG.................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TCCCCACTCTGCCAACAGCCACTGACACTC................................................................ | 30 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........TAAGTGGCCTGTGGAGTGATGGCACT...................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................TGTGGAGTGATGGCACTGTCTGTGCAAA........................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................TTGTAGATGGGTACACAGGCTTGGACA..................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TACATCTACCCACCAGCCATTGTGCAGC......... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................TAGATGGGTACACAGGCTTGGACAGGG.................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGAGGTGTTGCTGCATCTGTTTTGT............................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................CCACTGACACTCTGCTTGGCCCACAG.................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................GAGGTGTTGCTGCATCTGTTTTGTAGA......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TCTGTTTTGTAGATGGGTACACAGGC............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TCTGTTTTGTAGATGGGTACACAGGCT........................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TTGTAGATGGGTACACAGGCTTGGAC...................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGGCCTGTGGAGTGATGGCACTGTCT.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGTTGCTGCATCTGTTTTGTAGATG....................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................AGGTGTTGCTGCATCTGTTTTGTAGA......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TGGCGCTAAGTGGCCTGTGGAGTGATG........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................GTGTTGCTGCATCTGTTTTGTAGA......................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................ACTGTCTGTGCAAAGGCCCTGGGGTT............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................TGTTTTGTAGATGGGTACACAGGCTTt......................................................................................................................................... | 27 | t | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TCTGTGCAAAGGCCCTGGGGTTGGT............................................................................................................................................................................................ | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| GAGCTGGCGCTAAGTGGCCTGTGGAGTGATGGCACTGTCTGTGCAAAGGCCCTGGGGTTGGTGCCCACCTGAGGTGTTGCTGCATCTGTTTTGTAGATGGGTACACAGGCTTGGACAGGGCAGGGCACAGACCTATGCTCTCTGCACCCCTGGCCTTCCCCACTCTGCCAACAGCCACTGACACTCTGCTTGGCCCACAGGCTGACTCCTCACTACATCTACCCACCAGCCATTGTGCAGCCCAGCGTGG .....................................................................................(((((..(((((((((.....(((((.((...((((((((..((......))..)))))).)))).)))))..)))).))))).)))))............................................................................ .................................................................................82..........................................................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|