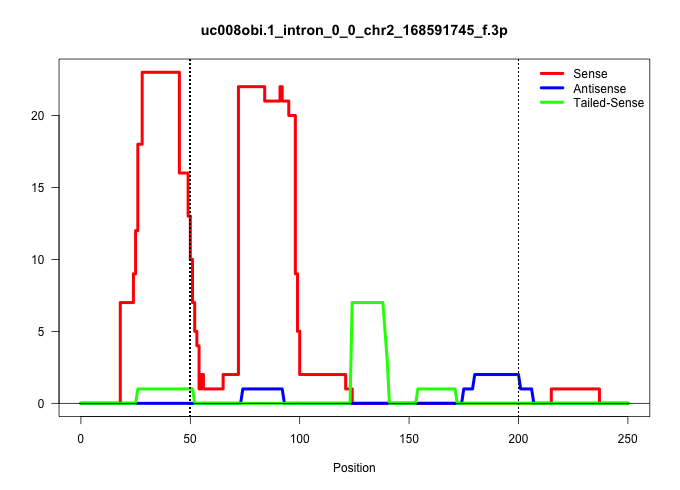
| Gene: AK052878 | ID: uc008obi.1_intron_0_0_chr2_168591745_f.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(3) PIWI.mut |
(17) TESTES |
| TATGTATCCAAAGGAGGGAAACCAATTATGGGACTGTGGACGAGGAGAGAGAAGTTGGAGGGAGAGGCAGAATGTGGAATCAACTTTACAATAAGAGGGAGAAAACTAGGGGGATTTGCAAAGGGGTGGGCGGGTTGAGGCTCCCTCCGAGGTGGGGGATTGAGGCTCCCTCCCTCCGCATCTCTCCTTTCCTGGAACAGAGTCCGGAATCCCCATTCCAGAATCGGCTCCCGGAACCCGCGCCCTCGAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................TGTGGAATCAACTTTACAATAAGAGG........................................................................................................................................................ | 26 | 1 | 11.00 | 11.00 | 6.00 | 2.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................AAACCAATTATGGGACTGTGGACGAGG............................................................................................................................................................................................................. | 27 | 1 | 7.00 | 7.00 | - | 6.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................TGTGGAATCAACTTTACAATAAGAGGGA...................................................................................................................................................... | 28 | 1 | 4.00 | 4.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGTGGAATCAACTTTACAATAAGAGGG....................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................GGTGGGCGGGTTGtaaa............................................................................................................. | 17 | taaa | 3.00 | 0.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TGGGACTGTGGACGAGGAGAGAGAAG.................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TATGGGACTGTGGACGAGGAGAG......................................................................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GGTGGGCGGGTTGtaa.............................................................................................................. | 16 | taa | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................GGTGGGCGGGTTGta............................................................................................................... | 15 | ta | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................TATGGGACTGTGGACGAGGAGAGAG....................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGTGGAATCAACTTTACAATAAG........................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................ATTATGGGACTGTGGACGAGGAGAGA........................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TTCCAGAATCGGCTCCCGGAAC............. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................TATGGGACTGTGGACGAGGAGAGA........................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................TATGGGACTGTGGACGAGGAGAGAaa...................................................................................................................................................................................................... | 26 | aa | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TTATGGGACTGTGGACGAGGAGAGAGA...................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TGGGACTGTGGACGAGGAGAGAGAAGTT.................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TTATGGGACTGTGGACGAGGAGAGA........................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................TTATGGGACTGTGGACGAGGAGAGAG....................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGGAGGGAGAGGCAGAATGTGGAATCAAC...................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................TAAGAGGGAGAAAACTAGGGGGATTTGCAA................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGCAGAATGTGGAATCAACTTTACAAT.............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GGGGATTGAGGCTCCagt.............................................................................. | 18 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................GAAAACTAGGGGGATTTGCAAAGG.............................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................TATGGGACTGTGGACGAGGAGAGAGA...................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TGGGACTGTGGACGAGGAGAGAGAA..................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................ATTATGGGACTGTGGACGAGGAGAG......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TATGTATCCAAAGGAGGGAAACCAATTATGGGACTGTGGACGAGGAGAGAGAAGTTGGAGGGAGAGGCAGAATGTGGAATCAACTTTACAATAAGAGGGAGAAAACTAGGGGGATTTGCAAAGGGGTGGGCGGGTTGAGGCTCCCTCCGAGGTGGGGGATTGAGGCTCCCTCCCTCCGCATCTCTCCTTTCCTGGAACAGAGTCCGGAATCCCCATTCCAGAATCGGCTCCCGGAACCCGCGCCCTCGAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................GGTGGGGGATTGAGagga...................................................................................... | 18 | agga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................TGGAATCAACTTTACAATA............................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCCTTTCCTGGAACAGAGTCCGG........................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................................................................CGGAACCCGCGtagg........ | 15 | tagg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CCGCATCTCTCCTTTCCTGGAACAGA................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................GAGTCCGGAATCgtc....................................... | 15 | gtc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |