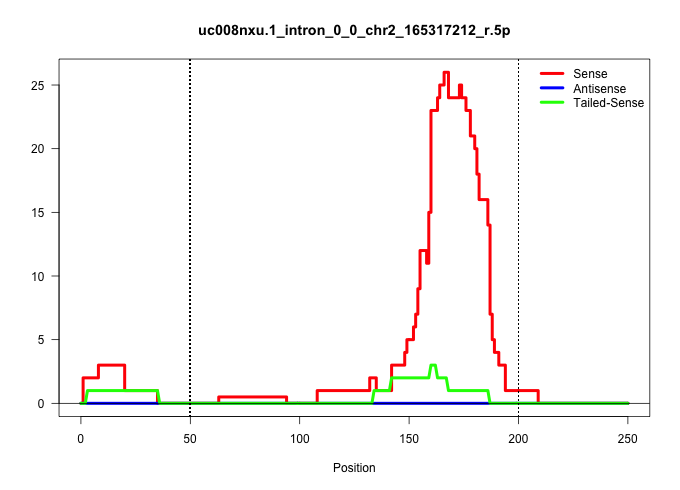
| Gene: 2810408M09Rik | ID: uc008nxu.1_intron_0_0_chr2_165317212_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(21) TESTES |
| GCGGCGGACAGTGCAGGAGGCGCGCGCGCTGCTCCGCTGCCGCCGTGCGGGTGAGGGGGGCCGGACGGGGCTCCGGGAGACGGGACAGACAGGGCTGCGGGAGACCAGTGGGACGTGAGAGACGGGGCTCCGATTTCTAGGGGACAGGATCAGATGGCAATGCTTAAGGACTCAAGGCGACAGTCAGGACGCCATTCTGTTTGGTTCGCGGGGCTTGTTTAGCGCTCGGGTCCCTCCGTTTCTCATTTCT ........................................................................................................((..(((((..((.(((......)))))..)))))..))......((...((((..(((((.........)))))...)))).))............................................................. .......................................................................................................104....................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TAAGGACTCAAGGCGACAG................................................................... | 19 | 1 | 11.00 | 11.00 | - | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGCTTAAGGACTCAAGGCGACAGTCAG............................................................... | 27 | 1 | 9.00 | 9.00 | - | - | 2.00 | 3.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................GTGGGACGTGAGAGACGG............................................................................................................................. | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...GCGGACAGTGCAGGAGGCGCGCGCG.............................................................................................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................ATGCTTAAGGACTCAAGGCGACAGTCAG............................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ...GCGGACAGTGCAGGAGGC..................................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................GACAGGATCAGATGGCAATGCTTAAG.................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GGCAATGCTTAAGGACTCAAGGCGACA.................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGCTTAAGGACTCAAGGCGACAGTCAGG.............................................................. | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TGGCAATGCTTAAGGACTCAAGGCGAC..................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .CGGCGGACAGTGCAGGAGG...................................................................................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GGCAATGCTTAAGGACTCAAGGCGACAGT.................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................ATCAGATGGCAATGCTTAAGGACTCA............................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................ATGCTTAAGGACTCAAGGCGACAGTCA................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TGGGACGTGAGAGACGGGGCTCCGATT................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CAGTGCAGGAGGCGCGCGCGCTGCTCC....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................GACAGGATCAGATGGCAATGCTTAAa.................................................................................. | 26 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................CAGGACGCCATTCTGTTTGGTTCGC......................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................TCAGATGGCAATGCTTAAGGACTCAAGGC........................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ATTTCTAGGGGACAGGATCAGATGGC............................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGCTTAAGGACTCAAGGCGACAGgcag............................................................... | 27 | gcag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................ATGCTTAAGGACTCAAG.......................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GCGCGCGCGCTGCggca...................................................................................................................................................................................................................... | 17 | ggca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................AGGACTCAAGGCGACAGTCAGGACG........................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGCTTAAGGACTCAAGGCGACAGTCA................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................TAAGGACTCAAGGCGACAGTCAGGA............................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................AAGGCGACAGTCAGGACGCCA........................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GATGGCAATGCTTAAGGACTCAAGGCGA...................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TTAAGGACTCAAGGCGACAGTCAGGACGCCA........................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................ATGGCAATGCTTAAGGACTCAAGGC........................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................TTCTAGGGGACAGGATCAGATGGCAATtt....................................................................................... | 29 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...GCGGACAGTGCAtggt....................................................................................................................................................................................................................................... | 16 | tggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GACGGGGCTCCGGGAGACGGGACAGACAGGG............................................................................................................................................................ | 31 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GCCGGACGGGGCTCCGGGAGACGGGACAGA................................................................................................................................................................. | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| GCGGCGGACAGTGCAGGAGGCGCGCGCGCTGCTCCGCTGCCGCCGTGCGGGTGAGGGGGGCCGGACGGGGCTCCGGGAGACGGGACAGACAGGGCTGCGGGAGACCAGTGGGACGTGAGAGACGGGGCTCCGATTTCTAGGGGACAGGATCAGATGGCAATGCTTAAGGACTCAAGGCGACAGTCAGGACGCCATTCTGTTTGGTTCGCGGGGCTTGTTTAGCGCTCGGGTCCCTCCGTTTCTCATTTCT ........................................................................................................((..(((((..((.(((......)))))..)))))..))......((...((((..(((((.........)))))...)))).))............................................................. .......................................................................................................104....................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................GGGCCGGACGGGGCTgga.................................................................................................................................................................................. | 18 | gga | 30.67 | 0.00 | 30.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GGGCCGGACGGGGCTCgga................................................................................................................................................................................. | 19 | gga | 25.00 | 0.00 | 24.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................GGGCCGGACGGGGCTCgg................................................................................................................................................................................. | 18 | gg | 5.00 | 0.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGGGACAGGATCAGATGG............................................................................................. | 19 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................GGCCGGACGGGGCTC................................................................................................................................................................................. | 15 | 2 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................GGCCGGACGGGGCTCtgg................................................................................................................................................................................. | 18 | tgg | 0.50 | 0.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GGGCCGGACGGGGCT.................................................................................................................................................................................. | 15 | 3 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |