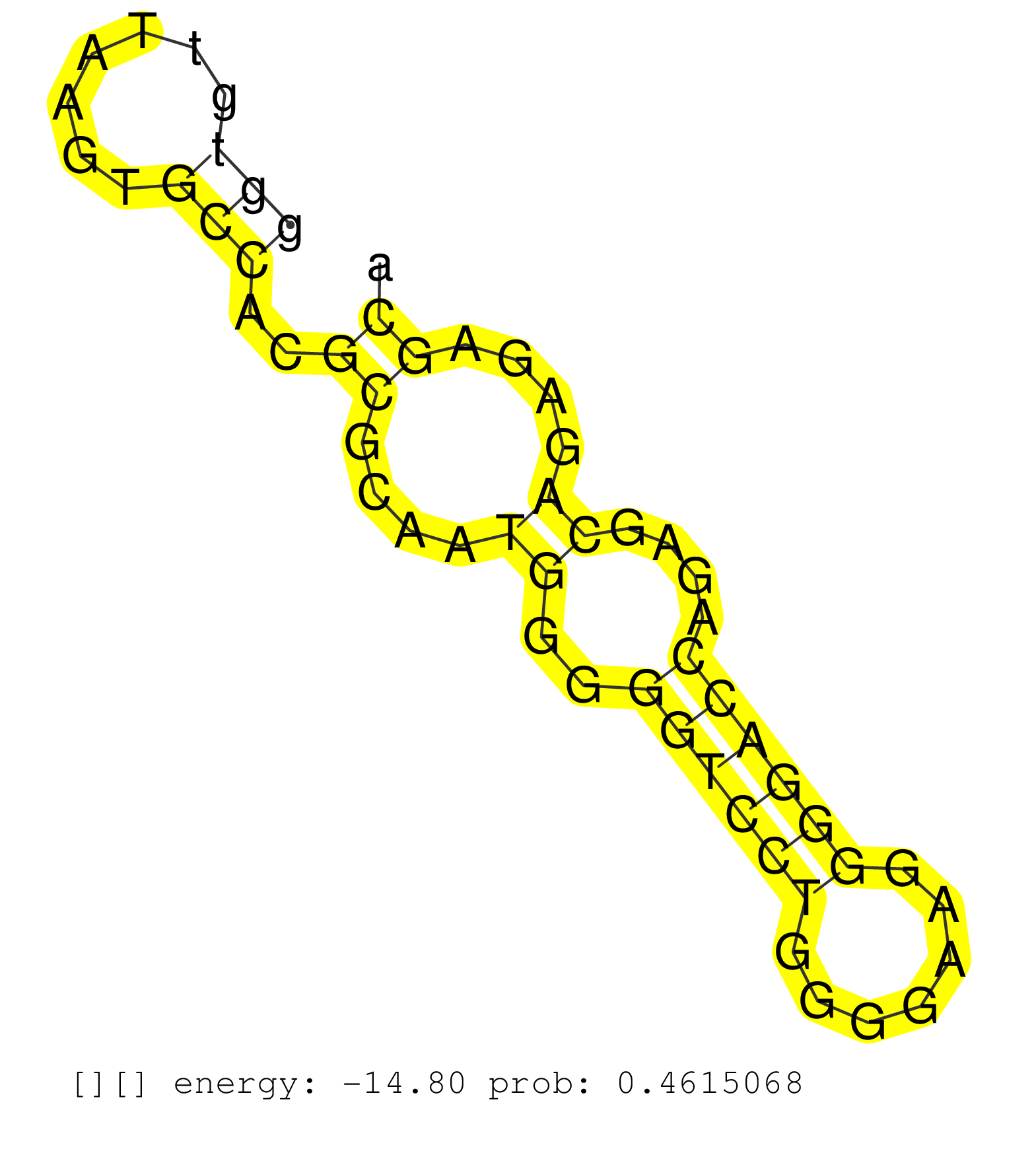
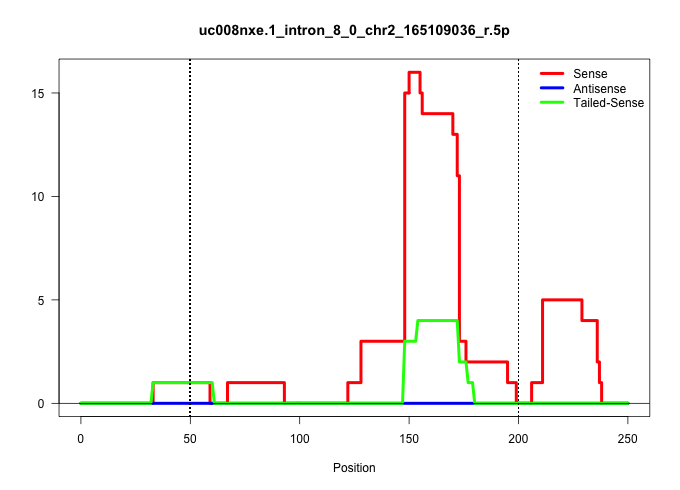
| Gene: Slc35c2 | ID: uc008nxe.1_intron_8_0_chr2_165109036_r.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(2) OVARY |
(4) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| CCCAGCTTTCCAGTGGCTCTGGGCCCGTTCGCCTGGCATTCGCCGAGCACGTACGAGGGGCCGGGTCTTGGGGATGGGGGGAACCAGGTGGATACTGCCGAGCAGGCGCGGCAGGGACACGTGAGCAAACACAGTCGTCACGCGGTGTTAAGTGCCACGCGCAATGGGGGTCCTGGGGAAGGGGACCAGAGCAGAGAGCATCAGGTTGCCTTGCCGAGGAGGACGCCGGCTGATGTTCCGGGCACACGGC ...............................................................................................................................................(((.......)))..((....((..((((((.......))))))....))....))................................................... ...............................................................................................................................................144.....................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................TAAGTGCCACGCGCAATGGGGGTCC............................................................................. | 25 | 1 | 9.00 | 9.00 | 5.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - |
| ....................................................................................................................................................TAAGTGCCACGCGCAATGGGGGTC.............................................................................. | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - |
| ...................................................................................................................................................................................................................TGCCGAGGAGGACGCCGGCTGATGT.............. | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAAGTGCCACGCGCAATGGGGGgcc............................................................................. | 25 | gcc | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAAGTGCCACGCGCAATGGGGGTCCTGaa......................................................................... | 29 | aa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TGGCATTCGCCGAGCACGTACGAGGG............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................AGTGCCACGCGCAATGGGGGTCCTGG.......................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAAGTGCCACGCGCAATGGGGGT............................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGGACGCCGGCTGATGTT............. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................ACACAGTCGTCACGCGGTGTTAAGTGCC.............................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGGGGAAGGGGACCAGAGCAGAGAGC................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................GAGCAAACACAGTCGTCACGCGGTGT...................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................TGGCATTCGCCGAGCACGTACGAGGGGt............................................................................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGGGGAAGGGGACCAGAGCAGAGAaa................................................... | 26 | aa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGGGGAAGGGGACCAGAGCAGAGAac................................................... | 26 | ac | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TGCCGAGGAGGACGCCGGCTGATGTT............. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................ACACAGTCGTCACGCGGTGTTAAGTGC............................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGTTAAGTGCCACGCGCAATGGGG................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................CCACGCGCAATGGGGGTCCTGGGGAt...................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGCCTTGCCGAGGAGGACGCCGG..................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TTGGGGATGGGGGGAACCAGGTGGAT............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GGGGACCAGAGCAGAGAG.................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................CCTGGGGAAGGGGACCAGAGCAGA....................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TGCCGAGGAGGACGCCGGCTGATGTTC............ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAAGTGCCACGCGCAATGGGGG................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCAGCTTTCCAGTGGCTCTGGGCCCGTTCGCCTGGCATTCGCCGAGCACGTACGAGGGGCCGGGTCTTGGGGATGGGGGGAACCAGGTGGATACTGCCGAGCAGGCGCGGCAGGGACACGTGAGCAAACACAGTCGTCACGCGGTGTTAAGTGCCACGCGCAATGGGGGTCCTGGGGAAGGGGACCAGAGCAGAGAGCATCAGGTTGCCTTGCCGAGGAGGACGCCGGCTGATGTTCCGGGCACACGGC ...............................................................................................................................................(((.......)))..((....((..((((((.......))))))....))....))................................................... ...............................................................................................................................................144.....................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................AACACAGTCGTCACGCGGT........................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |