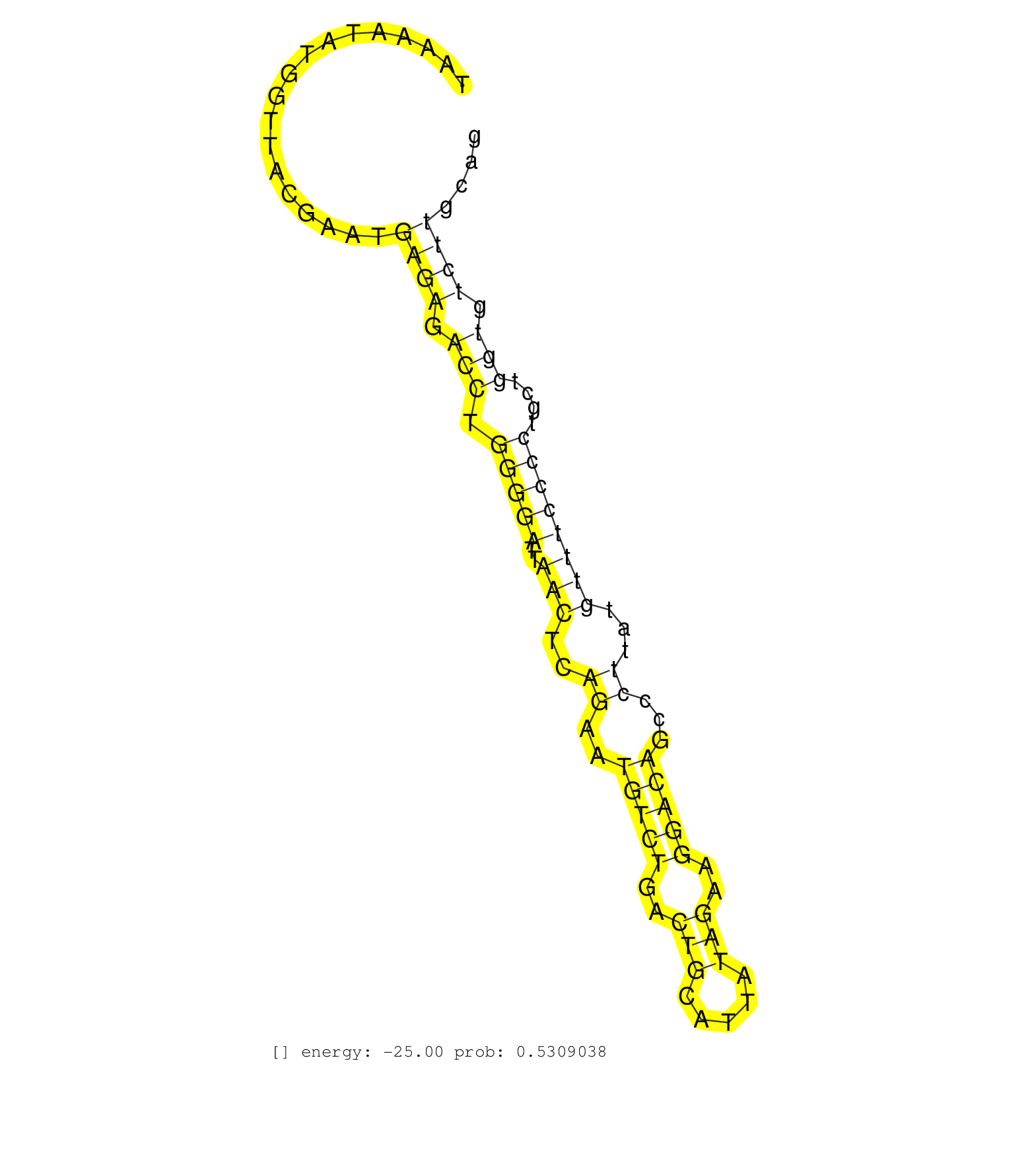

| Gene: Ncoa5 | ID: uc008nww.1_intron_3_0_chr2_164834950_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| TTCTTGGTCAGTTTAGTAGTAGAATTATATAAAGCATTACAGTGGCTGTGGCCTGTCACCCAAGGTTTTATCTTGATGGATTTAAGATAGTAGAAAAAAGTAAAATATGGTTACGAATGAGAGACCTGGGGATTAACTCAGAATGTCTGACTGCATTATAGAAGGACAGCCCTTATGTTTCCCCTGCTGGTGTCTTGCAGAGCGTTTGAAACGTGAGGAACGACGTAGAGAGGAGCTTTATCGTCGGTAT ......................................................................................................................((((.(((.(((((..(((..((..(((((..(((.....)))..)))))...))...))))))))....))).))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................TGATGGATTTAAGATAGTAGAAAAAAGTAAAATATGGTTACGAATGAGAGAC............................................................................................................................. | 52 | 1 | 87.00 | 87.00 | 87.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GAACGACGTAGAGAGGAG............... | 18 | 1 | 8.00 | 8.00 | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGAAACGTGAGGAACGACGTAGAGAG.................. | 26 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - |
| ..............................................................................................................................................................................................................TGAAACGTGAGGAACGACGTAGAGAGG................. | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGAAACGTGAGGAACGACGTAGAGA................... | 25 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGAAACGTGAGGAACGACGTAGAGAGGA................ | 28 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| .......................................................................................................................................................................................................................AGGAACGACGTAGAGAGGAGCTTTATC........ | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGAGGAACGACGTAGAGAGGAGCTTT........... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AACGTGAGGAACGACGTAGAGAGGAGCT............. | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGAAACGTGAGGAACGACGTAGAGAGGAt............... | 29 | t | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AACGTGAGGAACGACGTAGAGAGGA................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TAGAGAGGAGCTTTATCGTCGGT.. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................................TGAAACGTGAGGAACGACGTAGAGAGGtt............... | 29 | tt | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AACGTGAGGAACGACGTAGAGAGGAGC.............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AACGTGAGGAACGACGTAGAGAGGAGCTT............ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGAAACGTGAGGAACGACGTAGAGgat................. | 27 | gat | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GAAACGTGAGGAACGACGTAGAGAGGt................ | 27 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGAGGAACGACGTAGAGAGGAGCT............. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TTGAAACGTGAGGAACGACGTAGAG.................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................................................................................AAACGTGAGGAACGACGTAGAGAGGA................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| TTCTTGGTCAGTTTAGTAGTAGAATTATATAAAGCATTACAGTGGCTGTGGCCTGTCACCCAAGGTTTTATCTTGATGGATTTAAGATAGTAGAAAAAAGTAAAATATGGTTACGAATGAGAGACCTGGGGATTAACTCAGAATGTCTGACTGCATTATAGAAGGACAGCCCTTATGTTTCCCCTGCTGGTGTCTTGCAGAGCGTTTGAAACGTGAGGAACGACGTAGAGAGGAGCTTTATCGTCGGTAT ......................................................................................................................((((.(((.(((((..(((..((..(((((..(((.....)))..)))))...))...))))))))....))).))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|