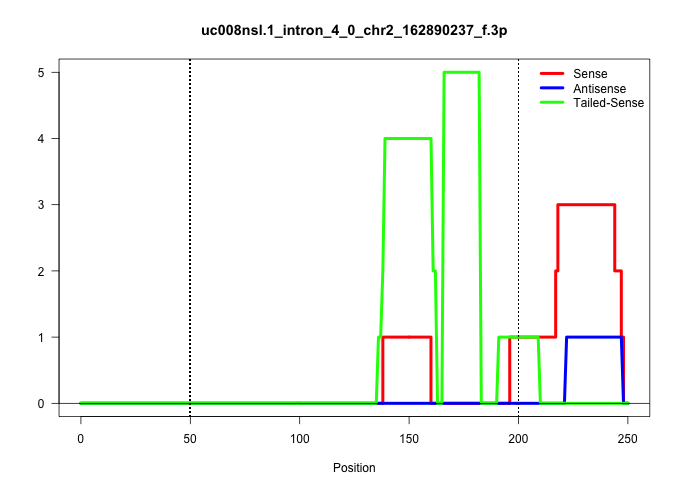
| Gene: Mybl2 | ID: uc008nsl.1_intron_4_0_chr2_162890237_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) OVARY |
(1) PIWI.ip |
(1) PIWI.mut |
(9) TESTES |
| TTCTCCATCCTGCCTTGAGTTCCACATTTGCTCATTTCATAGAGCTAATGAGGAAATTCACTAAACCTTCGAGGGCTGGAAACAGTGAGGCTTCACTGTACAGCTGGAGGATGCCTGTGTCCAGCCCTTCACGTTCAGGTGTGGGGAGAAGCTGCGGCAGCCGAGAGCTGCGTGTCAGAGGCCCCGGGTTTTCCCCACAGGACGGACAATGCTGTGAAGAATCACTGGAACTCCACTATCAAAAGGAAAG ......................................................................................................((((((((....))...))))))..............(((((((((((.(((.((..(((.....(((.....))).)))))))).)))))))))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................GCTGCGTGTCAGAcgtt................................................................... | 17 | cgtt | 5.00 | 0.00 | 5.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................AGAATCACTGGAACTCCACTATCAAAAGGA... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GTGTGGGGAGAAGCTGCGGCAG.......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................GAGGCCCCGGGTTTTt......................................................... | 16 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................AGGTGTGGGGAGAAGCTGCGGCAaa......................................................................................... | 25 | aa | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCCCCACAGGACGGACgga........................................ | 19 | gga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GAATCACTGGAACTCCACTATCAAAAGGAA.. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGTGGGGAGAAGCTGCGGCttt......................................................................................... | 22 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................TGTGGGGAGAAGCTGCGGCAGtat....................................................................................... | 24 | tat | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................GTGTGGGGAGAAGCTGCGGCAGCat....................................................................................... | 25 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| TTCTCCATCCTGCCTTGAGTTCCACATTTGCTCATTTCATAGAGCTAATGAGGAAATTCACTAAACCTTCGAGGGCTGGAAACAGTGAGGCTTCACTGTACAGCTGGAGGATGCCTGTGTCCAGCCCTTCACGTTCAGGTGTGGGGAGAAGCTGCGGCAGCCGAGAGCTGCGTGTCAGAGGCCCCGGGTTTTCCCCACAGGACGGACAATGCTGTGAAGAATCACTGGAACTCCACTATCAAAAGGAAAG ......................................................................................................((((((((....))...))))))..............(((((((((((.(((.((..(((.....(((.....))).)))))))).)))))))))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................TCACTGGAACTCCACTATCAAAAatc...... | 26 | atc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................ATTTGCTCATTTCAaatt................................................................................................................................................................................................................... | 18 | aatt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................................................................................CACTGGAACTCCACTATCAAAAGGAA.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |