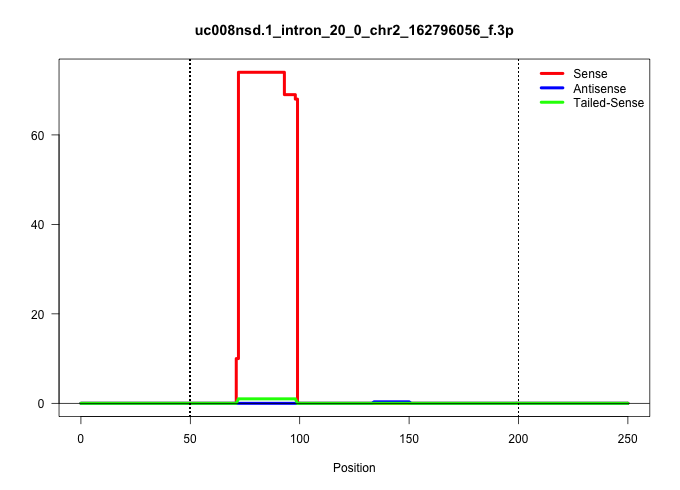
| Gene: L3mbtl | ID: uc008nsd.1_intron_20_0_chr2_162796056_f.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) PIWI.ip |
(17) TESTES |
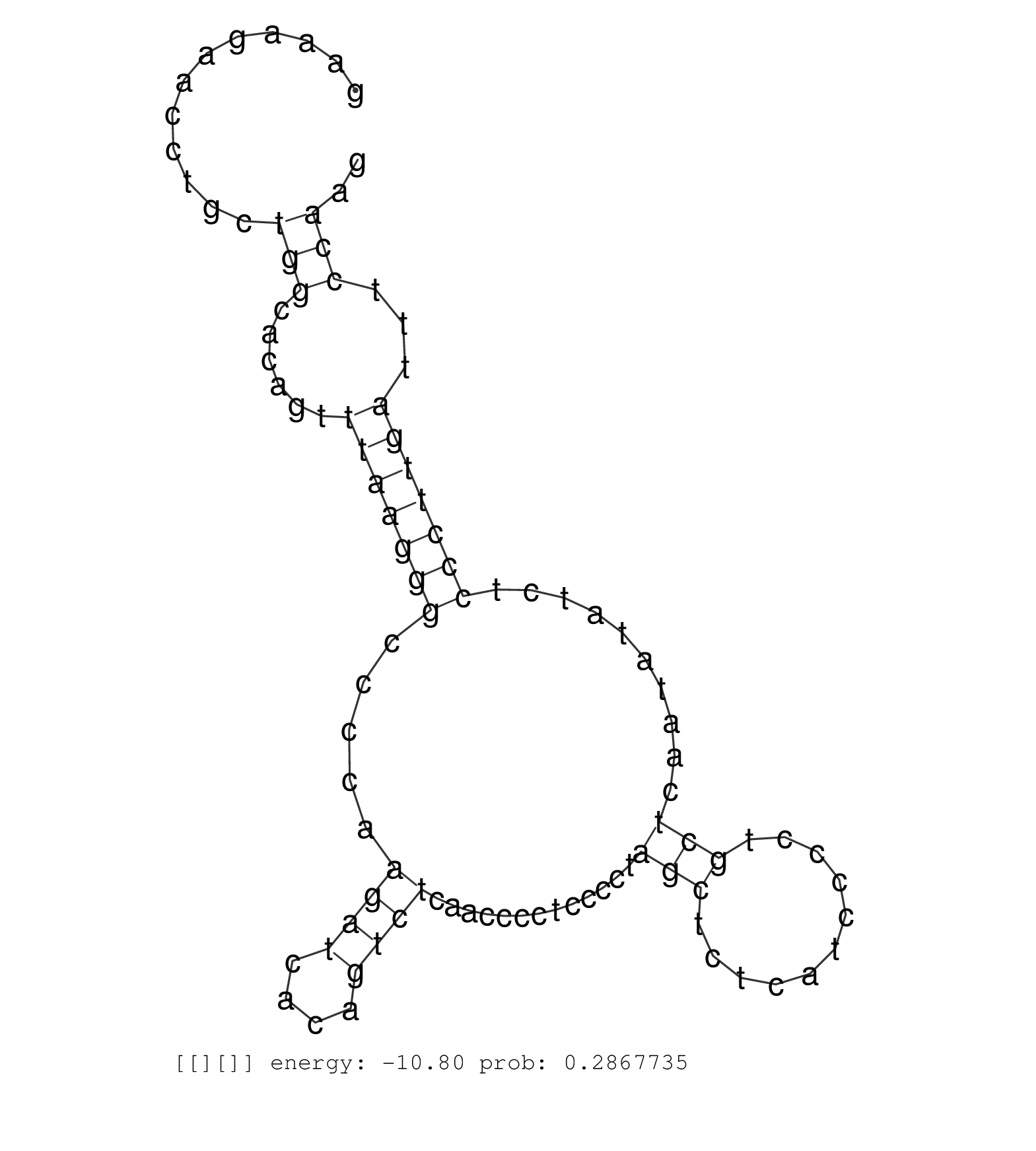
| CCTTGTAGATAGAGAGGGTGCTTCCTGTCTGCCATGCTTTAAGGAACAAGGGACCTTCCTTCATGTGCCTTAAGACATGATGATCCCCTGAGGCTGATAAGAAAGAACCTGCTGGCACAGTTTAAGGGCCCCAAGATCACAGTCTCAACCCCTCCCCTAGCTCTCATCCCCTGCTCAATATATCTCCCTTGATTTCCAAGGTCTTTGGCTTCGTTCAGACCTTGACGGGCTCTGAGGACCAAGCGCGCCT ................................................................................................................(((......(((((((.....((((....)))).............(((...........)))..........)))))))...))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................AGACATGATGATCCCCTGAGGCTGATA....................................................................................................................................................... | 27 | 1 | 68.00 | 68.00 | 38.00 | 9.00 | 5.00 | 4.00 | 3.00 | - | 2.00 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - |
| .......................................................................AAGACATGATGATCCCCTGAGGCTGATA....................................................................................................................................................... | 28 | 1 | 10.00 | 10.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 |
| ........................................................................AGACATGATGATCCCCTGAGG............................................................................................................................................................. | 21 | 1 | 5.00 | 5.00 | - | 1.00 | 1.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GACGGGCTCTGAGGAggc......... | 18 | ggc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGACATGATGATCCCCTGAGGCTGAT........................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGACATGATGATCCCCTGAGGCTGAca....................................................................................................................................................... | 27 | ca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCTTGTAGATAGAGAGGGTGCTTCCTGTCTGCCATGCTTTAAGGAACAAGGGACCTTCCTTCATGTGCCTTAAGACATGATGATCCCCTGAGGCTGATAAGAAAGAACCTGCTGGCACAGTTTAAGGGCCCCAAGATCACAGTCTCAACCCCTCCCCTAGCTCTCATCCCCTGCTCAATATATCTCCCTTGATTTCCAAGGTCTTTGGCTTCGTTCAGACCTTGACGGGCTCTGAGGACCAAGCGCGCCT ................................................................................................................(((......(((((((.....((((....)))).............(((...........)))..........)))))))...))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................GACATGATGATCCCCacgt.................................................................................................................................................................. | 19 | acgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................TCTCATCCCCTGCTCAtt......................................................................... | 18 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................GACATGATGATCCCCaaga.................................................................................................................................................................. | 19 | aaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................GATCACAGTCTCAACCC................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |