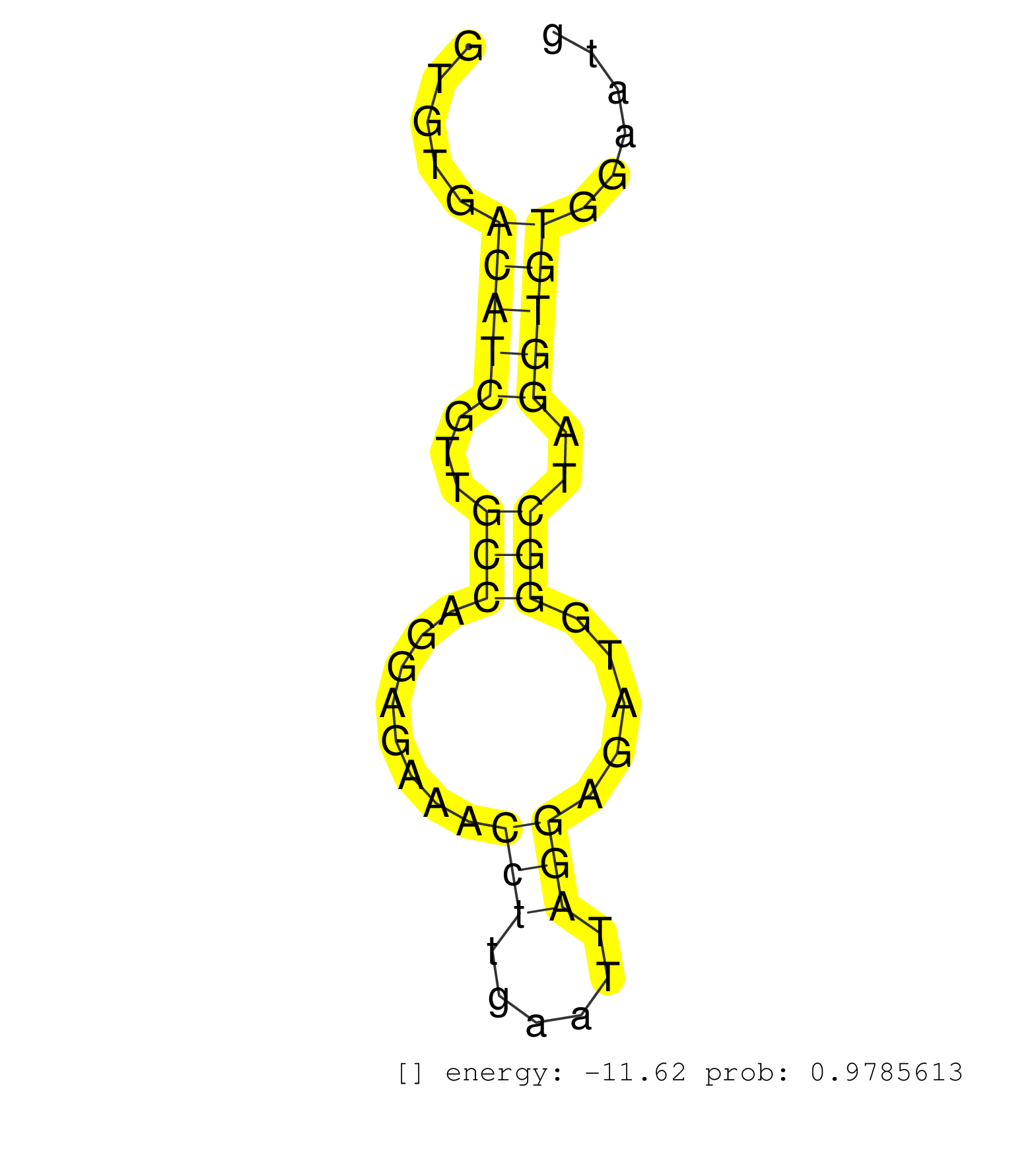

| Gene: Plcg1 | ID: uc008nrc.1_intron_1_0_chr2_160579090_f | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| CTGATGGCCTTCCTTCTCCCAACCAGCTCAGGAGGAAGATCCTTATTAAGGTGTGACATCGTTGCCAGGAGAAACCTTGAATTAGGAGATGGGCTAGGTGTGGAATGGTTTTTGCCATGACCTGGTCTGCTTCCACAGCACAAGAAGCTGGCTGAGGGCAGTGCCTATGAGGAGGTGCCTACCTCTGT .......................................................(((((...(((........(((......))).....)))..)))))....................................................................................... ..................................................51......................................................107............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................TTAGGAGATGGGCTAGGTGTGG..................................................................................... | 22 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 |
| .................................................................................TTAGGAGATGGGCTAGGTGTGt..................................................................................... | 22 | t | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGTGACATCGTTGCCAGGAGAAAC................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACAAGAAGCTGGCTGAGGGCAGTGCCT...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................GCACAAGAAGCTGGCTGAGGGCA............................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TAGGAGATGGGCTAGGTGTGGAATGG................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................ATTAGGAGATGGGCTAGGTGTGa..................................................................................... | 23 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................TTAGGAGATGGGCTAGGTGTGGAA................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGTGACATCGTTGCCAGGAGAAACC................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................AGCTCAGGAGGAAGATCCTTATTAAG.......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................AACCAGCTCAGGAGGAAGATCCTTATTAAG.......................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................AGGAGATGGGCTAGGTGTGGAAT.................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................TTAGGAGATGGGCTAGGTGTG...................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................ATTAGGAGATGGGCTAGGTGTGG..................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| CTGATGGCCTTCCTTCTCCCAACCAGCTCAGGAGGAAGATCCTTATTAAGGTGTGACATCGTTGCCAGGAGAAACCTTGAATTAGGAGATGGGCTAGGTGTGGAATGGTTTTTGCCATGACCTGGTCTGCTTCCACAGCACAAGAAGCTGGCTGAGGGCAGTGCCTATGAGGAGGTGCCTACCTCTGT .......................................................(((((...(((........(((......))).....)))..)))))....................................................................................... ..................................................51......................................................107............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................CCAACCAGCTCAGGAGGAAGATCCTTA............................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |