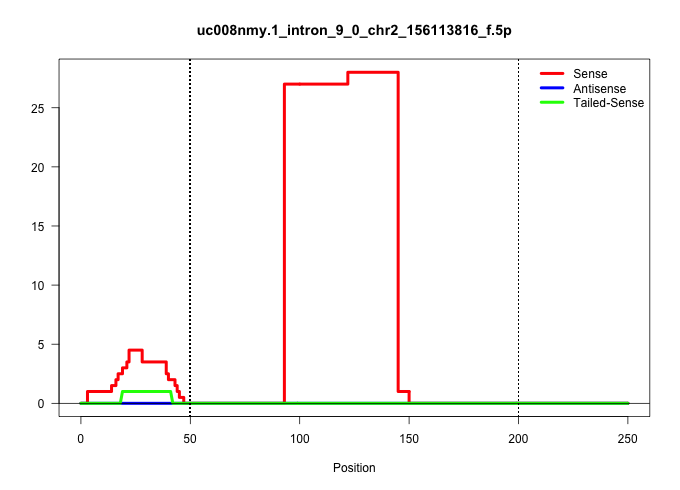
| Gene: Phf20 | ID: uc008nmy.1_intron_9_0_chr2_156113816_f.5p | SPECIES: mm9 |
 |
|
(2) PIWI.ip |
(9) TESTES |
| GACAAGCCAGGAGAGCCTAACCAGGAAGCGGGTCTCTGCCAGTTCCCCCAGTGAGTATCTTGGCTCTGTGGAATGTCCTAGGAGGGCCCTGGCATTTCTTAGGTGGAATGCTGCAGTAGGGTTGAGTAGTGACATTGGAGGACTGAAGCCCTCCTGGAAGAGGCAAGCTCGGCCTCTCTCTGTTCTTGTGGCAGTGAGGGAGTTACTGCTGAAGGCTGTGGCATTCCTGCGATGAGTAGGAGCTCTAAAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................ATTTCTTAGGTGGAATGCTGCAGTAGGGTTGAGTAGTGACATTGGAGGACTG......................................................................................................... | 52 | 1 | 27.00 | 27.00 | 27.00 | - | - | - | - | - | - | - | - |
| ...AAGCCAGGAGAGCCTAACCAGGAAG.............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ...................ACCAGGAAGCGGGTCTCTGCCAa................................................................................................................................................................................................................ | 23 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................TGAGTAGTGACATTGGAGGACTGAAGCC.................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ................CTAACCAGGAAGCGGGTCTCTGCCAGT............................................................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - |
| .................TAACCAGGAAGCGGGTCTCTGCCAGTTC............................................................................................................................................................................................................. | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - |
| .....................CAGGAAGCGGGTCTCTGCCAGTTCCC........................................................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - |
| ......................AGGAAGCGGGTCTCTGCC.................................................................................................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - |
| ..............GCCTAACCAGGAAGCGGGTCTCTGC................................................................................................................................................................................................................... | 25 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - |
| ......................AGGAAGCGGGTCTCTGCCAGTT.............................................................................................................................................................................................................. | 22 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - |
| ...................ACCAGGAAGCGGGTCTCTGC................................................................................................................................................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 |
| GACAAGCCAGGAGAGCCTAACCAGGAAGCGGGTCTCTGCCAGTTCCCCCAGTGAGTATCTTGGCTCTGTGGAATGTCCTAGGAGGGCCCTGGCATTTCTTAGGTGGAATGCTGCAGTAGGGTTGAGTAGTGACATTGGAGGACTGAAGCCCTCCTGGAAGAGGCAAGCTCGGCCTCTCTCTGTTCTTGTGGCAGTGAGGGAGTTACTGCTGAAGGCTGTGGCATTCCTGCGATGAGTAGGAGCTCTAAAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........AGCCTAACCAGGAAtcgg............................................................................................................................................................................................................................... | 18 | tcgg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - |