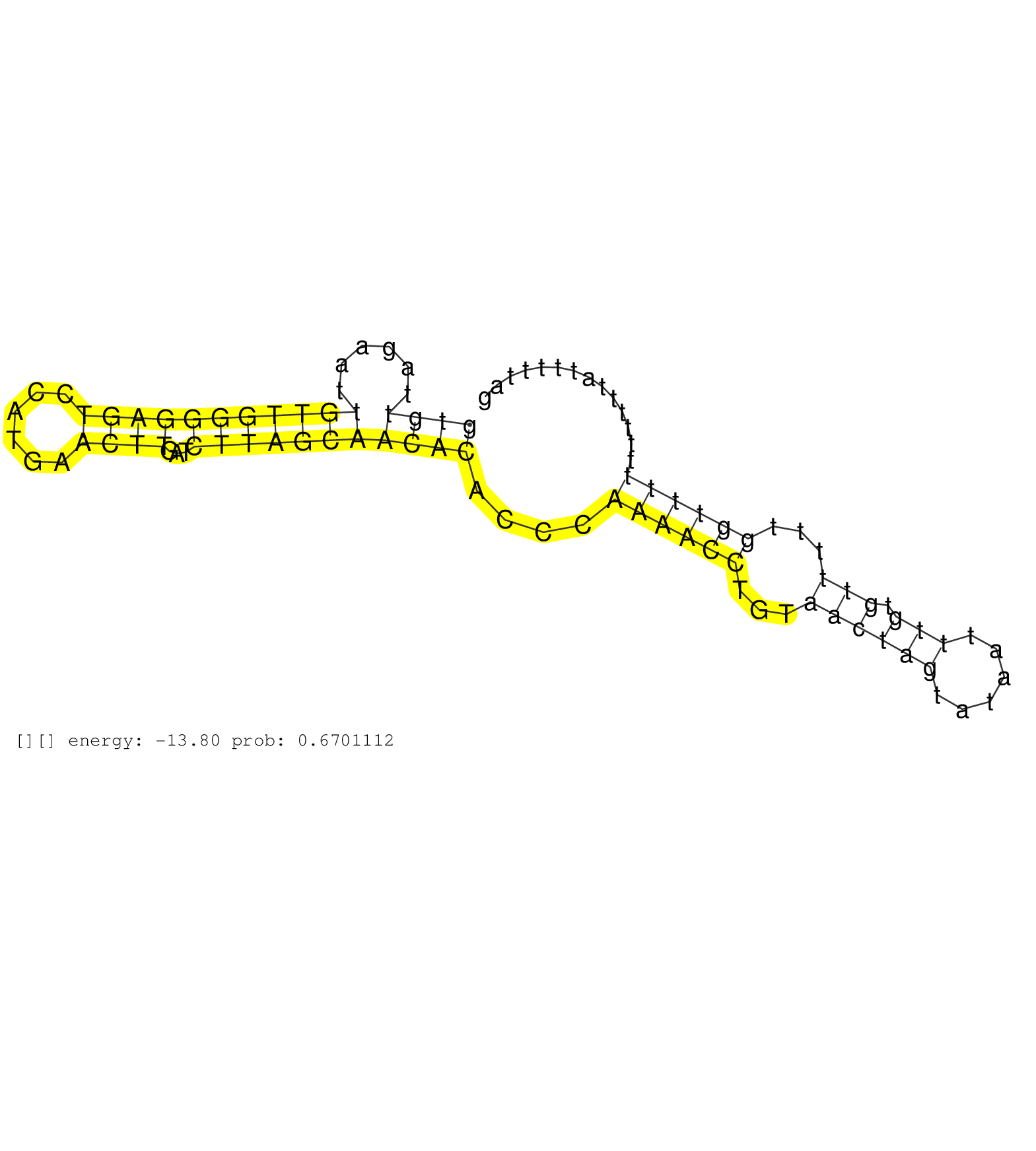

| Gene: Rbm39 | ID: uc008nmo.1_intron_12_0_chr2_155993487_r.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| GATAGAAAAACAAGGGCTACAGGTCTGGGATAAGTCATGATTTTTCTAAGTTCTGTTCTTTCCTCTTTATAGTTGAGGTAGAAAAGAAGCAGCATAAACAGTGTTAGAATTGTTGGGGAGTCCATGAACTTGATCTTAGCAACACACCCAAAACCTGTAACTAGTATAATTTGTGTTTTTGGTTTTTTTTTTATTTTTAGCTCCGGACCAAAATTTAACAGTGCCATCCGAGGAAAGATTGGATTGCCTC ....................................................................................................((((......(((((((((((......))))...)))))))))))....((((((...((((((......))).)))...))))))................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................GTTGGGGAGTCCATGAACTTGATCTTAGCAACACACCCAAAACCTGTAACTA....................................................................................... | 52 | 1 | 56.00 | 56.00 | 56.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................GTTGGGGAGTCCATGAACTTG...................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TTTAACAGTGCCATCCGAGGAAAGATT.......... | 27 | 2 | 1.50 | 1.50 | - | - | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .......................................................................................................................................................................................................................TAACAGTGCCATCCGAGGAAAGATTG......... | 26 | 2 | 1.50 | 1.50 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ........................................................................................................................................................................................................................AACAGTGCCATCCGAGGAAAGATTGGttt..... | 29 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................TTGAGGTAGAAAAGAAGCAGCATAAACAG..................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TTTAACAGTGCCATCCGAGGAAAGATg.......... | 27 | g | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................GCCATCCGAGGAAAGATTGGATTG.... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGGTCTGGGATAAGTCATGA.................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AAATTTAACAGTGCCATCCGAGGAAA.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................GAGGAAAGATTGGATTGCCT. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGTGCCATCCGAGGAAAGATTGGA....... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CTTTATAGTTGAGGTAGAAAAGAAGCAGCA............................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................TTTTAGCTCCGGACC......................................... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TTAACAGTGCCATCCGAGGAAAGATa.......... | 26 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TTAACAGTGCCATCCGAGGAAAGATTGGAg...... | 30 | g | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TTTATAGTTGAGGTAGAAAAGAAGCAGCATA.......................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................CTTTATAGTTGAGGTAGAAAAGAAGCAG.............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AATTTAACAGTGCCATCCGAGGAAAG............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................CATCCGAGGAAAGATTGGAT...... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........AAGGGCTACAGGTCTGGGATA.......................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCAAAATTTAACAGTGCCATCCGAGGA................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TAACAGTGCCATCCGAGGAAAGATTGGAT...... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................GTTAGAATTGTTGGGGAGTCCATGAAC......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CAGTGTTAGAATTGTTGGGGAGTCCATGAA.......................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGCCATCCGAGGAAAGAT........... | 18 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGTTAGAATTGTTGGG..................................................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GATAGAAAAACAAGGGCTACAGGTCTGGGATAAGTCATGATTTTTCTAAGTTCTGTTCTTTCCTCTTTATAGTTGAGGTAGAAAAGAAGCAGCATAAACAGTGTTAGAATTGTTGGGGAGTCCATGAACTTGATCTTAGCAACACACCCAAAACCTGTAACTAGTATAATTTGTGTTTTTGGTTTTTTTTTTATTTTTAGCTCCGGACCAAAATTTAACAGTGCCATCCGAGGAAAGATTGGATTGCCTC ....................................................................................................((((......(((((((((((......))))...)))))))))))....((((((...((((((......))).)))...))))))................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................TAGCTCCGGACgggt.......................................... | 15 | gggt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ACTTGATCTTAGCAACAgtag.......................................................................................................... | 21 | gtag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |