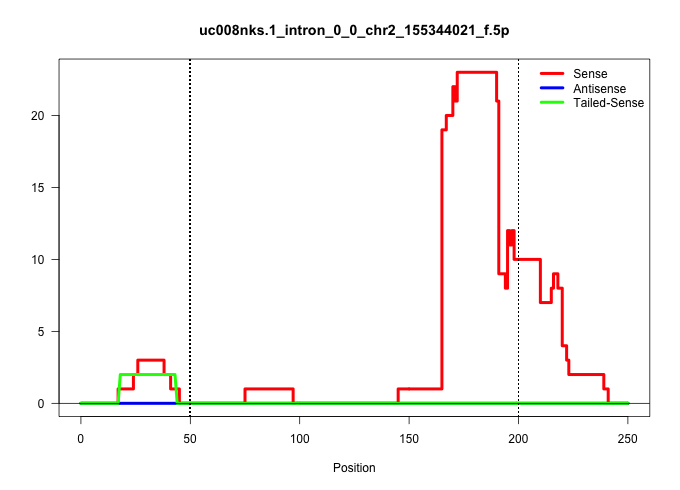
| Gene: Acss2 | ID: uc008nks.1_intron_0_0_chr2_155344021_f.5p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| TCTGCAGCGCTACCGCGAGCTGCACCGGCGTTCTGTGGAGGAGCCACGGGGTGAGGCTGGGCCCGAGCGACTCTTGGGGGCGCCAGTGAGCAGAGGAGCGGGAGTCCGAGGGAGATTTACCGAGCTCCGTTAGAAAGGGAGGAGATGGAGGGTCCCTGAGGGACATGGAAAAGGGATTCGAAAGAGATGGCGTTCTGTGAGAAGAAGGAGGCATCTGTGAGGCGAGACTTCCTAGAAGTTGGGGCACTCT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................TGGAAAAGGGATTCGAAAGAGATGGC........................................................... | 26 | 1 | 16.00 | 16.00 | 8.00 | - | 3.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................TGTGAGAAGAAGGAGGCATCTGTGA.............................. | 25 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................GTTCTGTGAGAAGAAGGAG........................................ | 19 | 2 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGGAAAAGGGATTCGAAAGAGATGG............................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - |
| ............................................................................................................................................................................GGGATTCGAAAGAGATGGCGTTCTGT.................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................GCTGCACCGGCGTTCTGTGGAGGAGt.............................................................................................................................................................................................................. | 26 | t | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................AAGGGATTCGAAAGAGATGGCGTTCT...................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TGAGAAGAAGGAGGCATCTGTGAGG............................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CCGGCGTTCTGTGGAGG................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................AAGGGATTCGAAAGAGATGGCGTTC....................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................GGCGTTCTGTGGAGGAGCC............................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................GGGGGCGCCAGTGAGCAGAGGA......................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................GTTCTGTGAGAAGAAGGAGGCATCTGT................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGTGAGAAGAAGGAGGCATCTGTGAGGC........................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGTGAGGCGAGACTTCCTAGAAGTTG......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................GAAAAGGGATTCGAAAGAGATGGCGTT........................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGAGGGTCCCTGAGGGACATGGAAA............................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GTGAGGCGAGACTTCCTAGAAGT........... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................AGCTGCACCGGCGTTCTGTGG.................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| TCTGCAGCGCTACCGCGAGCTGCACCGGCGTTCTGTGGAGGAGCCACGGGGTGAGGCTGGGCCCGAGCGACTCTTGGGGGCGCCAGTGAGCAGAGGAGCGGGAGTCCGAGGGAGATTTACCGAGCTCCGTTAGAAAGGGAGGAGATGGAGGGTCCCTGAGGGACATGGAAAAGGGATTCGAAAGAGATGGCGTTCTGTGAGAAGAAGGAGGCATCTGTGAGGCGAGACTTCCTAGAAGTTGGGGCACTCT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|