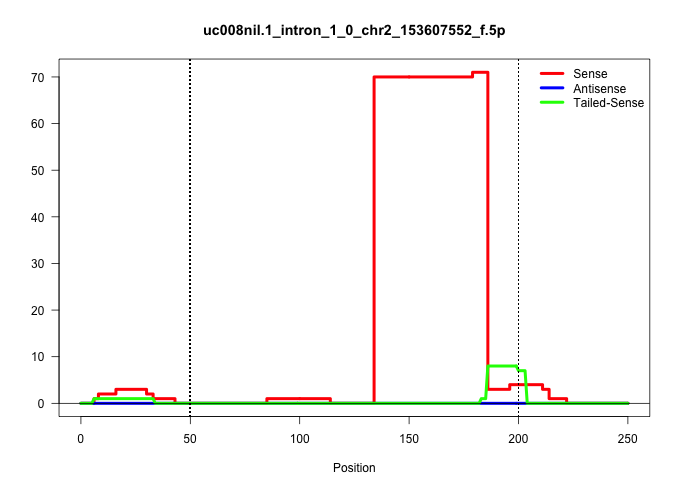
| Gene: EG329541 | ID: uc008nil.1_intron_1_0_chr2_153607552_f.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(11) TESTES |
| CAAGACTAATGTCTTCTGAGAAGCCAGGAGAGAGCCCCAAACCTCAAAAGGTAAGAACATAACAGTGAGTGAGGGGAGATCACAATGGGAGCCACACAAATGTCATTTACCAGTCCACTGCCTTCCAAAATGTCACCAGCATAGGTCTGCTAGCTTCTGCACCAGGCTTGCAAACTTAGATATGTGTAGGACCAAGTGATAACTTAGACGAGCCTAACAAGCAGGTTCCTCTTCCCACCCCTCCTCTCCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................ACCAGCATAGGTCTGCTAGCTTCTGCACCAGGCTTGCAAACTTAGATATGTG................................................................ | 52 | 1 | 70.00 | 70.00 | 70.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TAGGACCAAGTGATAgca.............................................. | 18 | gca | 7.00 | 0.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TAGGACCAAGTGATAACTTAGACGAGCC.................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ......TAATGTCTTCTGAGAAGCCAGGAGAGAa........................................................................................................................................................................................................................ | 28 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................TGATAACTTAGACGAGCCTAACAAGC............................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......TAATGTCTTCTGAGAAGCCAGGAG............................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................GTGTAGGACCAAGgccg.................................................. | 17 | gccg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................TGAGAAGCCAGGAGAGAGCCCCAAACC............................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................ATATGTGTAGGACCAAGTGATAACTTAGACGA....................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........ATGTCTTCTGAGAAGCCAGGAGAGA......................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................TGGGAGCCACACAAATGTCATTTACCAGT........................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| CAAGACTAATGTCTTCTGAGAAGCCAGGAGAGAGCCCCAAACCTCAAAAGGTAAGAACATAACAGTGAGTGAGGGGAGATCACAATGGGAGCCACACAAATGTCATTTACCAGTCCACTGCCTTCCAAAATGTCACCAGCATAGGTCTGCTAGCTTCTGCACCAGGCTTGCAAACTTAGATATGTGTAGGACCAAGTGATAACTTAGACGAGCCTAACAAGCAGGTTCCTCTTCCCACCCCTCCTCTCCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................CCCCAAACCTCAAAgatt.......................................................................................................................................................................................................... | 18 | gatt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |