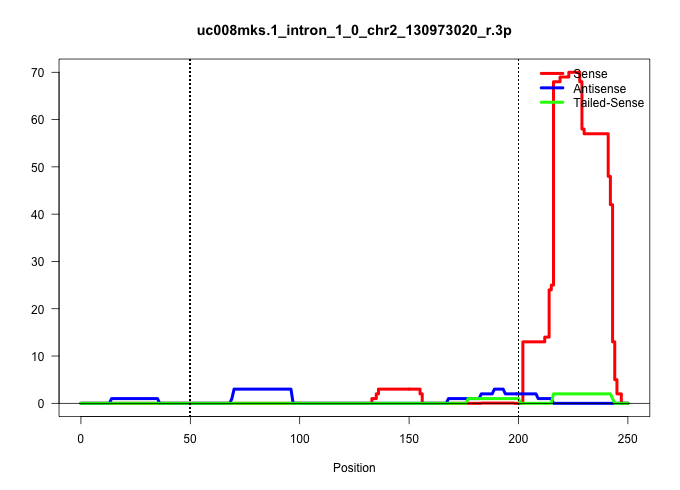
| Gene: 1700037H04Rik | ID: uc008mks.1_intron_1_0_chr2_130973020_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(5) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(23) TESTES |
| CTTCCACCTTTAGAGCCACACTGAGTTCTCAGGCCAGGGGCACATTGGCCTTATTTGAAAGGATGGGGAGTTTCTTAAGTCCTGTACCAGCTCTGTAGGGTGGGTCTCTGAAGGGACCATCTGGGGACTGTGCATGCTGGGGACAGTATGGAGGAGCTGTAGGAGATACTGATTGCCTATCCATCCCTGTCCTCCAACAGGTTACAGCATCAAGTGTGAGTACTCGGCACACAAAGAAGGTGTTCTCAAA ......................................................................................................................................((.(((((((((.(((((...((..(((......)))...))...)))))..)))))).))).))................................................... ................................................................................................................................129....................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................TGAGTACTCGGCACACAAAGAAGGTGT....... | 27 | 1 | 25.00 | 25.00 | 8.00 | 5.00 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - |
| ..........................................................................................................................................................................................................TACAGCATCAAGTGTGAGTACTCGGCA..................... | 27 | 1 | 10.00 | 10.00 | 6.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGTACTCGGCACACAAAGAAGGTGTT...... | 28 | 1 | 8.00 | 8.00 | 1.00 | 3.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGTGAGTACTCGGCACACAAAGAAGGT......... | 27 | 1 | 5.00 | 5.00 | - | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGTACTCGGCACACAAAGAAGGTG........ | 26 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................................................TGTGAGTACTCGGCACACAAAGAAGGTGT....... | 29 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGTACTCGGCACACAAAGAAGGTGTTC..... | 29 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGTACTCGGCACACAAAGAAGGT......... | 25 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGCTGGGGACAGTATGGAGGAGC............................................................................................. | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGTGAGTACTCGGCACACAAAGAAGGTG........ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGTGAGTACTCGGCACACAAAGAAGG.......... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CTGGGGACAGTATGGAGGAGCTaaga........................................................................................ | 26 | aaga | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TACAGCATCAAGTGTGAGTACTCGGC...................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................TATCCATCCCTGTCCTCCAACAGt................................................. | 24 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................ATGCTGGGGACAGTATGGAGGAGC............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CTGGGGACAGTATGGAGGAGC............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................ATGCTGGGGACAGTATGGAGGA............................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GCTGGGGACAGTATGGAGGAG.............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TACAGCATCAAGTGTGAGTACTCGGCAC.................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGTACTCGGCACACAAAGAAGGTGct...... | 28 | ct | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGTACTCGGCACACAAAGAAGGTGc....... | 27 | c | 1.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GTACTCGGCACACAAAGAAGGTGTTCTC... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TCGGCACACAAAGAAGGTGTTCTC... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GTGAGTACTCGGCACACAAAGAAGGT......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CTGGGGACAGTATGGAGGAG.............................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................AGTGTGAGTACTCGGCACACAAAGAAGGTGT....... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TCCCTGTCCTCCAAC.................................................... | 15 | 18 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - |
| CTTCCACCTTTAGAGCCACACTGAGTTCTCAGGCCAGGGGCACATTGGCCTTATTTGAAAGGATGGGGAGTTTCTTAAGTCCTGTACCAGCTCTGTAGGGTGGGTCTCTGAAGGGACCATCTGGGGACTGTGCATGCTGGGGACAGTATGGAGGAGCTGTAGGAGATACTGATTGCCTATCCATCCCTGTCCTCCAACAGGTTACAGCATCAAGTGTGAGTACTCGGCACACAAAGAAGGTGTTCTCAAA ......................................................................................................................................((.(((((((((.(((((...((..(((......)))...))...)))))..)))))).))).))................................................... ................................................................................................................................129....................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................TTTCTTAAGTCCTGTACCAGCTCTGTA......................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TCCCTGTCCTCCAACAGGTTACAGCA......................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................GGAGTTTCTTAAGTCCTGTACCAGCTCT............................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GTTTCTTAAGTCCTGTACCAGCTCTGTA......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............GCCACACTGAGTTCTCAGGCCA...................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................TCCTCCAACAGGTTACAGCATCAAGTG.................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CTGATTGCCTATCCATCCCTGTCCTC........................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |