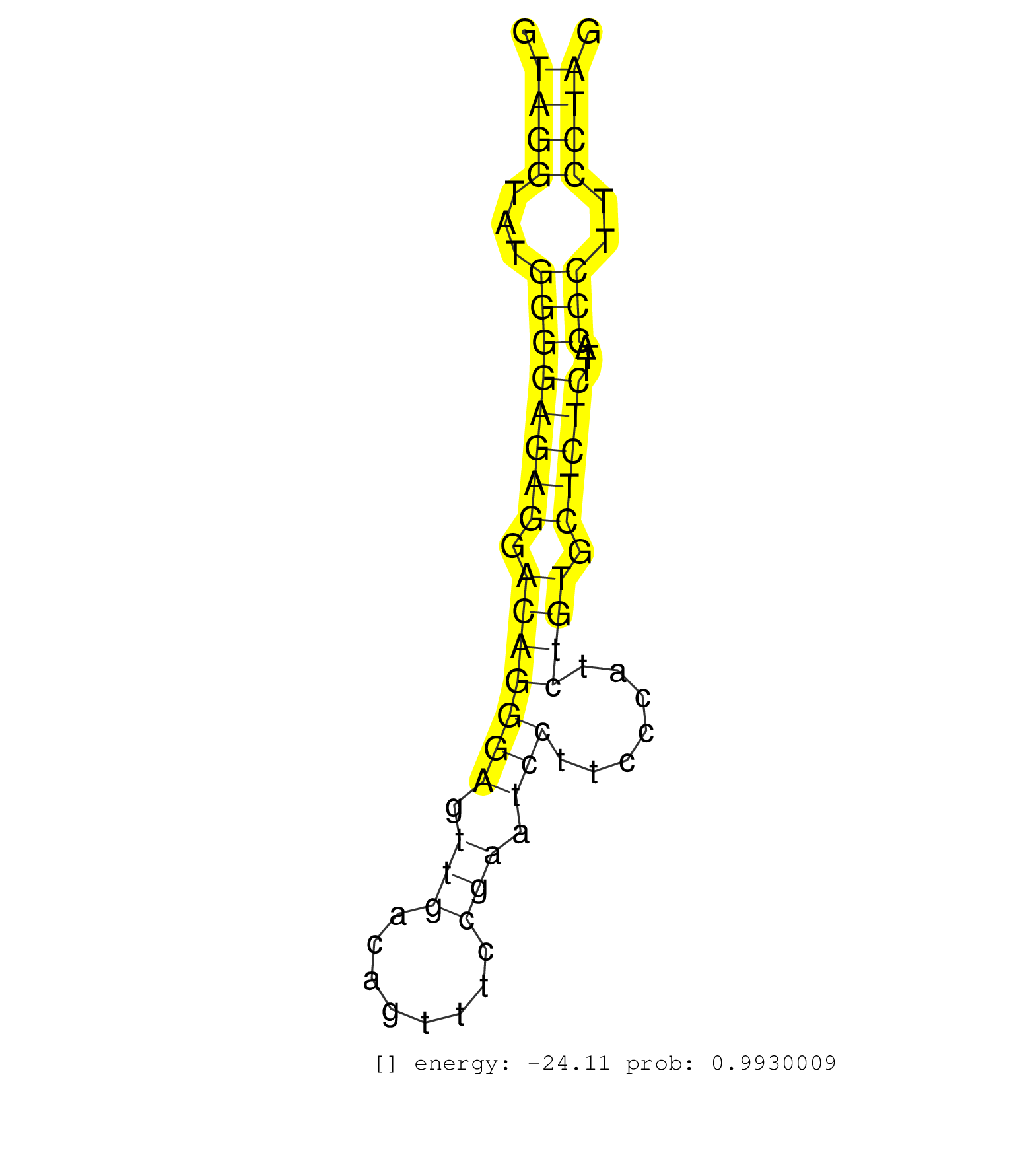
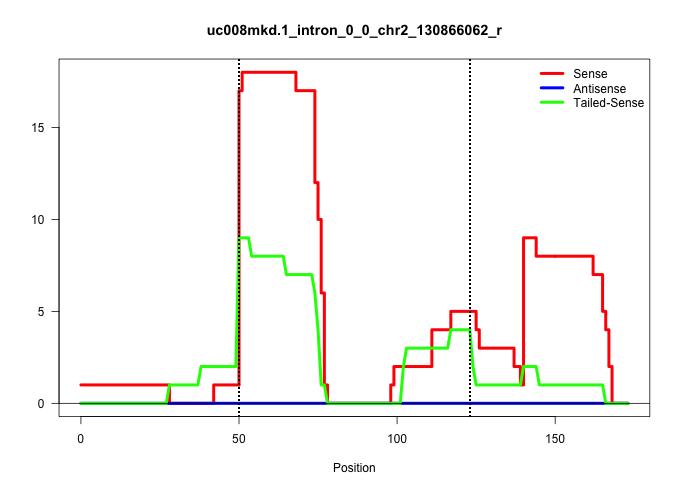
| Gene: Gfra4 | ID: uc008mkd.1_intron_0_0_chr2_130866062_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| TGCAGCCATCAGTTCTGCAGGACCAGACTGCTGGGTGCTGTTTCCCGCGGGTAGGTATGGGGAGAGGACAGGGAGTTGACAGTTTCCGAATCCTTCCCATCTGTGCTCTCTTACCCTTCCTAGGCAAGGCACGAGTGGCCTGAGAAGAGCTGGAGGCAGAAACAGTCCTTGTT ...................................................((((...((((((((.(((((((.(((........))).))).......)))).)))))...)))..))))................................................... ..................................................51......................................................................123................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGTATGGGGAGAGGACAGGGA................................................................................................... | 24 | 1 | 5.00 | 5.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAAGAGCTGGAGGCAGAAACAGTC...... | 27 | 1 | 5.00 | 5.00 | - | 1.00 | 1.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATGGGGAGAGGACAGGGAGTT................................................................................................ | 27 | 1 | 4.00 | 4.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATGGGGAGAGGACAGGGAGT................................................................................................. | 26 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAAGAGCTGGAGGCAGAAACAGTCC..... | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAAGAGCTGGAGGCAGAAACAG........ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - |
| ......................................................................................................GTGCTCTCTTACCCTTCCTAGt................................................. | 22 | t | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATGGGGAGAGGACAGGGAG.................................................................................................. | 25 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATGGGGAGAGGACAGGGAta................................................................................................. | 26 | ta | 2.00 | 5.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATGGGGAGAGGACAGGGAGTTG............................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTAGGTATGGGGAGAGGACAGGGAa.................................................................................................. | 25 | a | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATGGGGAGAGGACAGGGAGa................................................................................................. | 26 | a | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TCCTAGGCAAGGCACGAGTGGCCTGAG............................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................TACCCTTCCTAGGCAAGGCACGAGTGGC.................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TCTGTGCTCTCTTACCCTTCCTAGGC................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TCCCGCGGGTAGGTATGGGGAGAGGA......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGCTCTCTTACCCTTCCTAGtt................................................ | 22 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAAGAGCTGGAGGCAGAAAtagt....... | 26 | tagt | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................TGCTGGGTGCTGTTTCCCGCGGGTAt....................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................TGAGAAGAGCTGGAGGCAGAAACAGT....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TCCTAGGCAAGGCACGAGTGGCCTGAGc............................ | 28 | c | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................ATCTGTGCTCTCTTACCCTTCCTAGGCA............................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................TGAGAAGAGCTGGAGGCAGAAA........... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATGGGGAGAGGACAGGGAGTTt............................................................................................... | 28 | t | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATGGGGAGAGGACAGGGt................................................................................................... | 24 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACCCTTCCTAGGCAAGGCACGAGTG.................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAGGTATGGGGAGAGGACAGGGAt.................................................................................................. | 25 | t | 1.00 | 5.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TGTTTCCCGCGGGTAGGTATGGGGAGg............................................................................................................ | 27 | g | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGTATGGGGAGAGGACAGGGAGTT................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGCAGCCATCAGTTCTGCAGGACCAGACTGCTGGGTGCTGTTTCCCGCGGGTAGGTATGGGGAGAGGACAGGGAGTTGACAGTTTCCGAATCCTTCCCATCTGTGCTCTCTTACCCTTCCTAGGCAAGGCACGAGTGGCCTGAGAAGAGCTGGAGGCAGAAACAGTCCTTGTT ...................................................((((...((((((((.(((((((.(((........))).))).......)))).)))))...)))..))))................................................... ..................................................51......................................................................123................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|