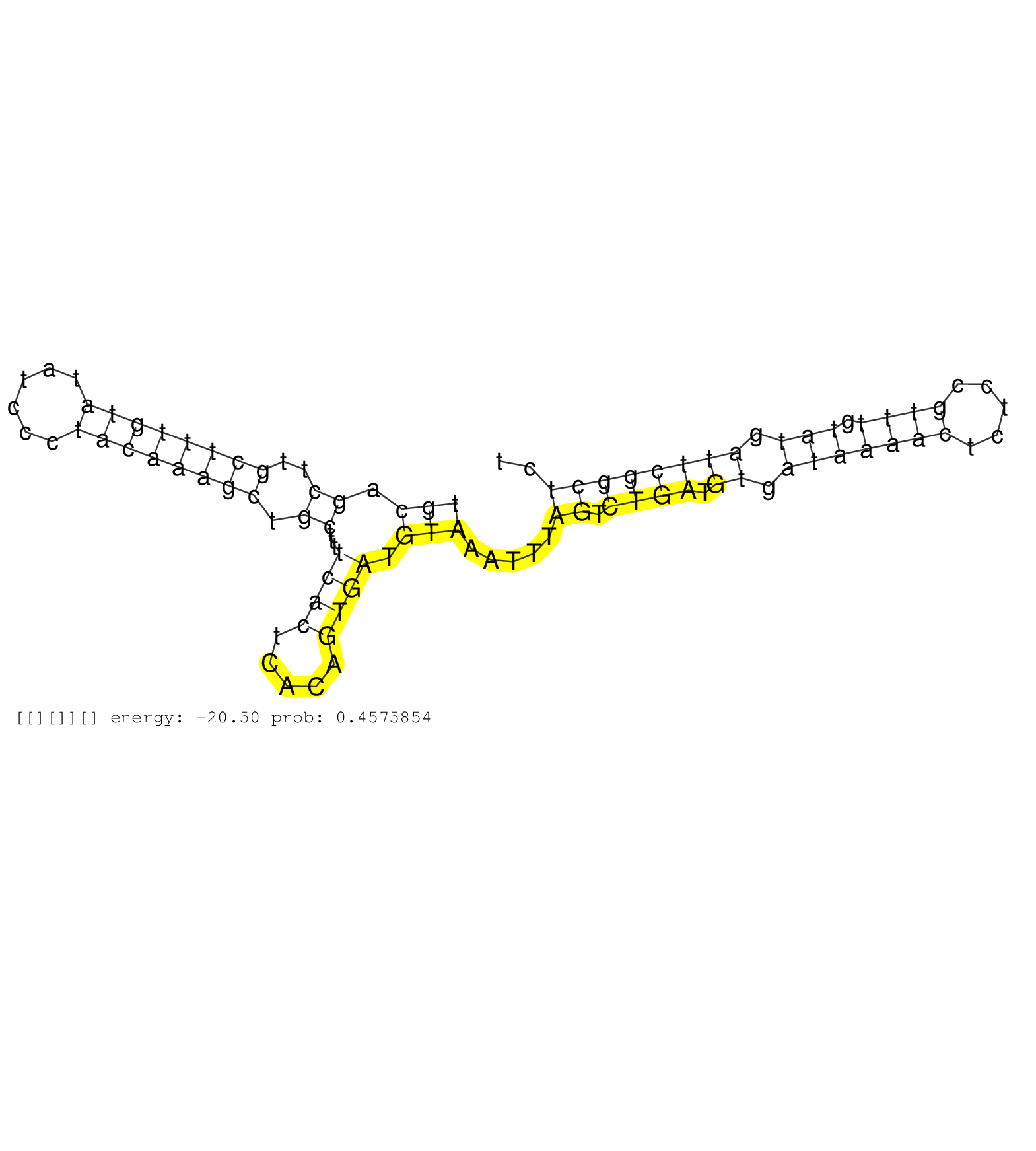

| Gene: Atrn | ID: uc008mkc.1_intron_24_0_chr2_130839405_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| CAGCACAGCAACTTCATGGACCTGGTACAGTTCTTCGTGACTTTCTTCAGGTAATTTCTCTATGCTAATTGTACACATTCCATCGAGACAGTCCCTTAACTGCAGCTTGCTTTGTATATCCCTACAAAGCTGCTTTTCACTCACAGTGATGTAAATTTAGTCTGATGTGATAAAACTCTCCGTTTGTATGATTCGGCTCTTTGCATGGGGAGAGGTTTGGGCTCAAGCAGTTATTAATAATATAGCTACT ....................................................................................................(((.((..((((((((......)))))))).))...((((.....)))).))).....((.((((.((.(((((((.....)))).))).)))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................CACAGTGATGTAAATTTAGTCTGATG................................................................................... | 26 | 1 | 37.00 | 37.00 | 10.00 | 10.00 | 6.00 | 4.00 | 2.00 | 1.00 | 2.00 | - | 1.00 | 1.00 | - | - | - |
| ............................................................................................................................................TCACAGTGATGTAAATTTAGTCTGATG................................................................................... | 27 | 1 | 9.00 | 9.00 | 3.00 | - | - | 1.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - |
| ................TGGACCTGGTACAGTTCTTCGTGACTTT.............................................................................................................................................................................................................. | 28 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - |
| .............................................................................................................................................CACAGTGATGTAAATTTAGTCTGATGTG................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCACAGTGATGTAAATTTAGTCTG...................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TGGACCTGGTACAGTTCTTCGTGACTTTC............................................................................................................................................................................................................. | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...............ATGGACCTGGTACAGTTCTTCGTGACTTT.............................................................................................................................................................................................................. | 29 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| .......................GGTACAGTTCTTCGTGAC................................................................................................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ..........ACTTCATGGACCTGGT................................................................................................................................................................................................................................ | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - |
| CAGCACAGCAACTTCATGGACCTGGTACAGTTCTTCGTGACTTTCTTCAGGTAATTTCTCTATGCTAATTGTACACATTCCATCGAGACAGTCCCTTAACTGCAGCTTGCTTTGTATATCCCTACAAAGCTGCTTTTCACTCACAGTGATGTAAATTTAGTCTGATGTGATAAAACTCTCCGTTTGTATGATTCGGCTCTTTGCATGGGGAGAGGTTTGGGCTCAAGCAGTTATTAATAATATAGCTACT ....................................................................................................(((.((..((((((((......)))))))).))...((((.....)))).))).....((.((((.((.(((((((.....)))).))).)))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................CCTGGTACAGTTCTTCGTGACTTTCTTCA......................................................................................................................................................................................................... | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |