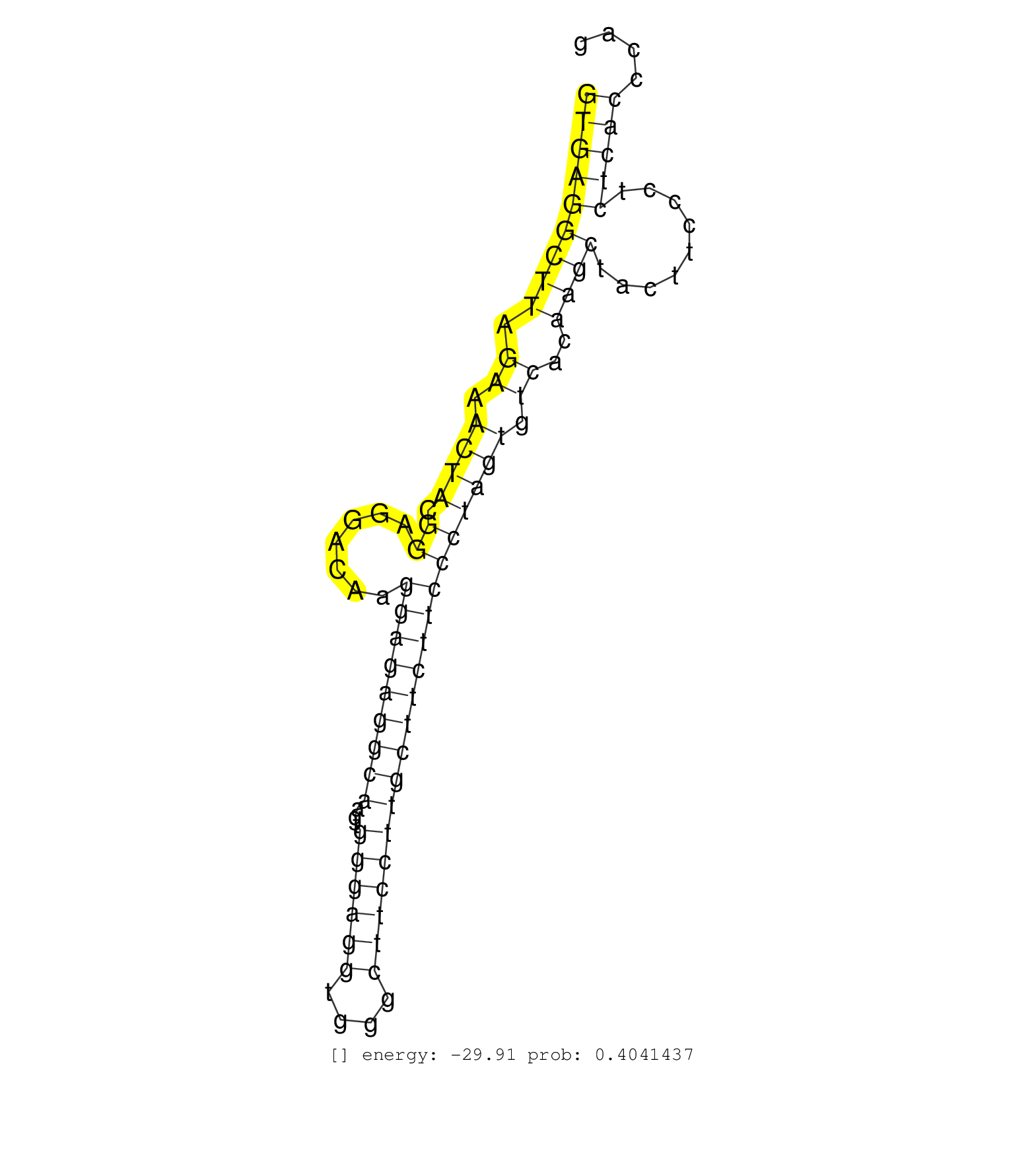

| Gene: Idh3b | ID: uc008min.1_intron_5_0_chr2_130107277_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(3) PIWI.mut |
(20) TESTES |
| TTATCATTCGAGAGCAGACAGAAGGGGAGTATAGCTCTCTGGAACATGAGGTGAGGCTTAGAAACTACGGAGGACAAGGAGAGGCAAGTGGGAGGTGGGCTTCCTTGCTTCTTCCCTAGTGTCACAAGCTACTTCCCTCTCACCCAGAGCGCCAAGGGTGTCATTGAGTGCCTGAAGATCGTCACTCGCACCAAGTC ..................................................(((((((((.((.((((.((.......(((((((((...((((((....))))))))))))))))))))).))..)))).........)))))...................................................... ..................................................51..............................................................................................147................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGGCTTAGAAACTACGGAGGACA......................................................................................................................... | 26 | 1 | 31.00 | 31.00 | 3.00 | 10.00 | 2.00 | 3.00 | 6.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 |
| ..................................................GTGAGGCTTAGAAACTACGGAGGACAA........................................................................................................................ | 27 | 1 | 9.00 | 9.00 | 1.00 | - | 3.00 | 2.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGCTTAGAAACTACGGAGGACAAG....................................................................................................................... | 28 | 1 | 4.00 | 4.00 | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................AGTGCCTGAAGATCGTC.............. | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................AGGGTGTCATTGAGTGCCTGAAGATCGT............... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGCTTAGAAACTACGGAGGACAAG....................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGCTTAGAAACTACGGAGGACAAGG...................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGCTTAGAAACTACGGAGGACAA........................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TTCGAGAGCAGACAGAAGGGGAGTA...................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TATCATTCGAGAGCAGACAGAAGGGGAa........................................................................................................................................................................ | 28 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGCTTAGAAACTACGGAGGACAAGGA..................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................CTGAAGATCGTCACTCGCAC...... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................TGAGTGCCTGAAGATCGTCACTC.......... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................TGTCATTGAGTGCCTGAAGATCGTCAC............ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................GCCTGAAGATCGTCACTCGCACCAAGT. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................TGAGGCTTAGAAACTACGGAGGACAAGG...................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGCTTAGAAACTACGGAGGAC.......................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AAACTACGGAGGACAAGGAGAGGC................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGCTTAGAAACTACGGAGGACt......................................................................................................................... | 26 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TTCGAGAGCAGACAGAAGGGGAGTATt.................................................................................................................................................................... | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTATCATTCGAGAGCAGACAGAAGGGGAGTATAGCTCTCTGGAACATGAGGTGAGGCTTAGAAACTACGGAGGACAAGGAGAGGCAAGTGGGAGGTGGGCTTCCTTGCTTCTTCCCTAGTGTCACAAGCTACTTCCCTCTCACCCAGAGCGCCAAGGGTGTCATTGAGTGCCTGAAGATCGTCACTCGCACCAAGTC ..................................................(((((((((.((.((((.((.......(((((((((...((((((....))))))))))))))))))))).))..)))).........)))))...................................................... ..................................................51..............................................................................................147................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................CTACTTCCCTCTCACaatt...................................................... | 19 | aatt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TCCTTGCTTCTTCCCTAGTGTCACAAG..................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................GCCTGAAGATCGTCtact.............. | 18 | tact | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |