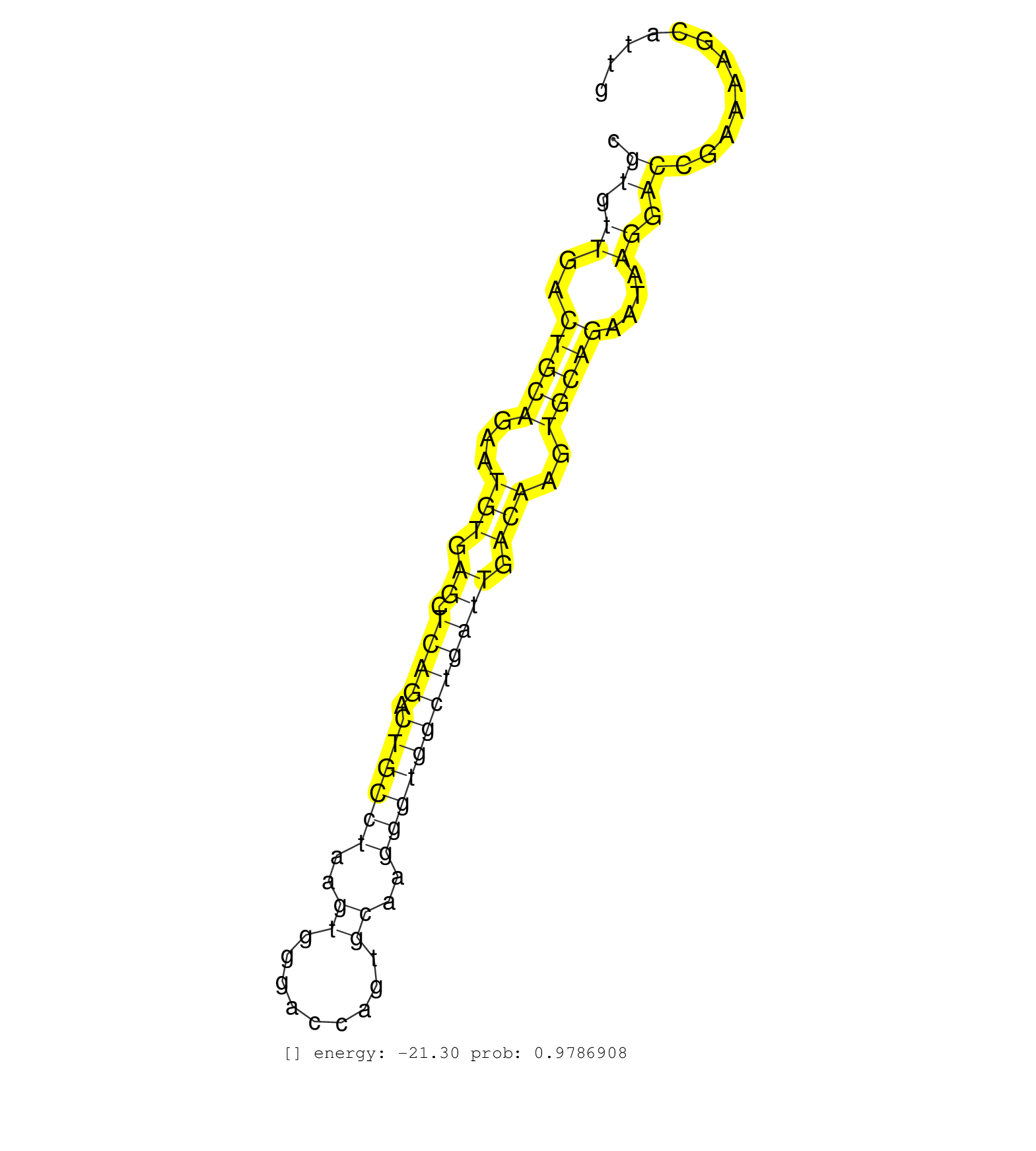

| Gene: Mertk | ID: uc008mgt.1_intron_8_0_chr2_128597237_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(12) TESTES |
| GGGCATCGGGCCCTTCAGTGAGCCAGTGAATATCATCATTCCTGAACACAGTGAGTGGGGCGGGGCGTGTTGACTGCAGAATGTGAGCTCAGACTGCCTAAGTGGGACCAGTGCAAGGGTGGCTGATTGACAAGTGCAGAATAAGGACCGAAAAGCATTGTGCAAGAACTCCAACACTCCTGCCATTACTGAAGATCAGTCCTTTCCAAATACTGAATTGGGCTCTACCTGTTGAATTTGGTTTAAGGCC ..................................................................((.((..(((((...(((.((.((((.((((((..((.........))..)))))))))))).)))..)))))....)).))...................................................................................................... .................................................................66............................................................................................160........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................TGACTGCAGAATGTGAGCTCAGACTGC......................................................................................................................................................... | 27 | 1 | 9.00 | 9.00 | - | 2.00 | 1.00 | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - |
| ........................................................................................................................................................................................................................ATTGGGCTCTACCTGTTGAAT............. | 21 | 1 | 8.00 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGATTGACAAGTGCAGAATAAGGACC..................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................ATGTGAGCTCAGACTGCCTAAGTGG................................................................................................................................................. | 25 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - |
| ......................................................................TGACTGCAGAATGTGAGCTCAGACT........................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGACAAGTGCAGAATAAGGACCGAAAAGC.............................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TAAGGACCGAAAAGCATTGTGCAAGAAC................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........CCTTCAGTGAGCCAGTGAATATC........................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................AATAAGGACCGAAAAGCATTGTGCA...................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................TGCAGAATGTGAGCTCAGACTGCCTA...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................TTGACTGCAGAATGTGAGCTCAGACTGC......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGACAAGTGCAGAATAAGGACCGAAAA................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................TCATTCCTGAACACAGTGAGTGGGGCGG........................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGGGGCGGGGCGTGggga................................................................................................................................................................................. | 19 | ggga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................GATTGACAAGTGCAGAATAAGGACCGAAAAGC.............................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................TAAGTGGGACCAGTGCAAGGGTGGCTG............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCAGACTGCCTAAGTGGGACCAGTGC........................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................TGACTGCAGAATGTGAGCTCAGACTGt......................................................................................................................................................... | 27 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGTGAGCTCAGACTGCCTAAGTGGG................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................AAGGACCGAAAAGCATTGTGCAAGAA.................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| GGGCATCGGGCCCTTCAGTGAGCCAGTGAATATCATCATTCCTGAACACAGTGAGTGGGGCGGGGCGTGTTGACTGCAGAATGTGAGCTCAGACTGCCTAAGTGGGACCAGTGCAAGGGTGGCTGATTGACAAGTGCAGAATAAGGACCGAAAAGCATTGTGCAAGAACTCCAACACTCCTGCCATTACTGAAGATCAGTCCTTTCCAAATACTGAATTGGGCTCTACCTGTTGAATTTGGTTTAAGGCC ..................................................................((.((..(((((...(((.((.((((.((((((..((.........))..)))))))))))).)))..)))))....)).))...................................................................................................... .................................................................66............................................................................................160........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................GGACCAGTGCAAggga...................................................................................................................................... | 16 | ggga | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |