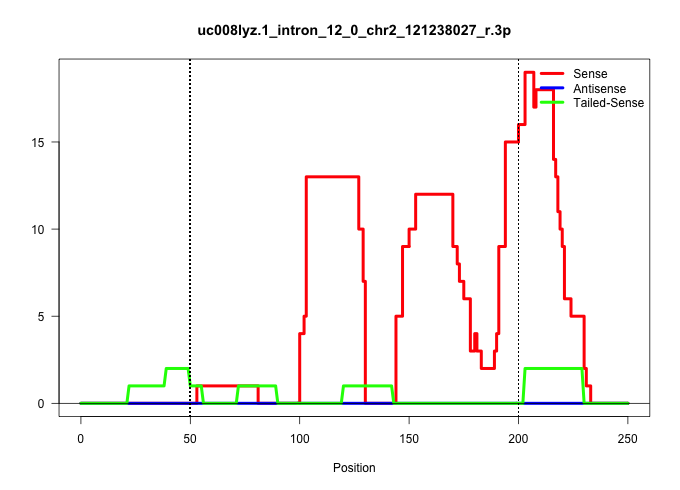
| Gene: Catsper2 | ID: uc008lyz.1_intron_12_0_chr2_121238027_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(19) TESTES |
| CTTAGATGAAAAACAAAATCCGTACCGAACAACAACAAAATACCTTTGAGTCTTTCAATGTGTTTATTCTTAACTGCTCATAGCTTTTTTTGTGTGGGCTTCTTGAATCTCTAAAAGGCAAAGAAGCTGGGGGAGGAGAGAGCCTGCTAATTGTAGTATAATTCTTAAGCATGACATCCTTGGTTCTACTTTCTTAACAGACATGGCACAAGAACAAGGACATTTCCAGCTGCTCAGAGCTGATGCTATC ....................................................................................................((((...(((((....(((......))).)))))...))))..((((.....)))).............................................................................................. ..................................................................................................99.........................................................................174.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................TGAATCTCTAAAAGGCAAAGAAGCTGG........................................................................................................................ | 27 | 1 | 6.00 | 6.00 | - | 3.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGAATCTCTAAAAGGCAAAGAAGCTG......................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TCTTGAATCTCTAAAAGGCAAAGAAGC........................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................TGCTAATTGTAGTATAATTCTTAAGC................................................................................ | 26 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGCACAAGAACAAGGACATTTCCAGC.................... | 27 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGAATCTCTAAAAGGCAAAGAAGCTGGGG...................................................................................................................... | 29 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAATTGTAGTATAATTCTTAAGCATGACATC........................................................................ | 31 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................TAACAGACATGGCACAAGAACAAGGAC............................. | 27 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCTTAACAGACATGGCACAAGAACA.................................. | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ACATGGCACAAGAACAAGGACATTTCCAGC.................... | 30 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................TAACAGACATGGCACAAGAACAAGGA.............................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................TACCGAACAACAACAAAATACCTTgagt........................................................................................................................................................................................................ | 28 | gagt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TTGAATCTCTAAAAGGCAAAGAAGCTG......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTTCTTAACAGACATGGCACAAGAACA.................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCTAATTGTAGTATAATTCTTAAGCAT.............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAATTGTAGTATAATTCTTAAGCATGAC........................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ATGGCACAAGAACAAGta.............................. | 18 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................CAAGAACAAGGACATTTCCAGCTGC................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCTAATTGTAGTATAATTCTTAAGCATG............................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGCACAAGAACAAGGACATTTCCAtc.................... | 27 | tc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGCACAAGAACAAGGACATTTCCAGCT................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TTGAATCTCTAAAAGGCAAAGAAGCTGG........................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................ATACCTTTGAGTCgtcg.................................................................................................................................................................................................. | 17 | gtcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................ACTGCTCATAGCTTcgtt................................................................................................................................................................ | 18 | cgtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................TCTTAACAGACATGGCACAAGAACAA................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCTTAACAGACATGGCACAAGAACAAG................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TAGTATAATTCTTAAGCATGACATCC....................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGCACAAGAACAAGGACATTTCtagc.................... | 27 | tagc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCTTAACAGACATGGCACAAGAACAAGG............................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TAACAGACATGGCACAAGAACAAGGACATT.......................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TAACAGACATGGCACAAGAACAAG................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AAGAAGCTGGGGGAGGAGAGAaa........................................................................................................... | 23 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGGTTCTACTTTCTTAACAGACATGGC........................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TCTTGAATCTCTAAAAGGCAAAGAAGCTGG........................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TTCAATGTGTTTATTCTTAACTGCTCAT......................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................TTGTAGTATAATTCTTAAGCATGACATCCTT..................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTGGTTCTACTTTCTTAACAGACATGGC........................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TAGTATAATTCTTAAGCATGACATCCTTGG................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TTCTTAACAGACATGGCACAAGAACA.................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTTAGATGAAAAACAAAATCCGTACCGAACAACAACAAAATACCTTTGAGTCTTTCAATGTGTTTATTCTTAACTGCTCATAGCTTTTTTTGTGTGGGCTTCTTGAATCTCTAAAAGGCAAAGAAGCTGGGGGAGGAGAGAGCCTGCTAATTGTAGTATAATTCTTAAGCATGACATCCTTGGTTCTACTTTCTTAACAGACATGGCACAAGAACAAGGACATTTCCAGCTGCTCAGAGCTGATGCTATC ....................................................................................................((((...(((((....(((......))).)))))...))))..((((.....)))).............................................................................................. ..................................................................................................99.........................................................................174.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|