
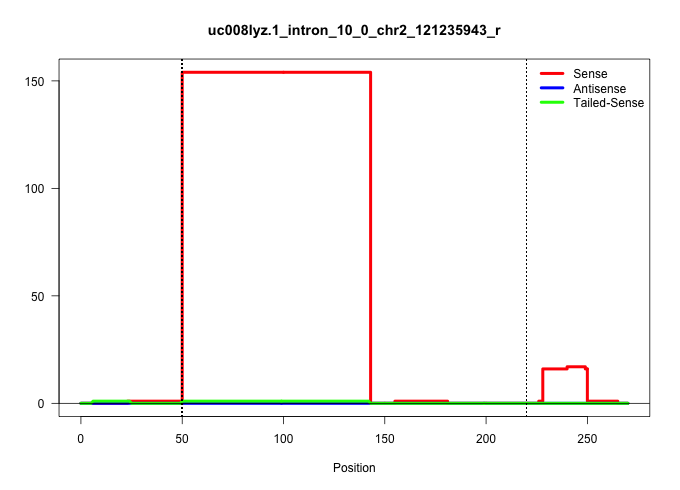
| Gene: Catsper2 | ID: uc008lyz.1_intron_10_0_chr2_121235943_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(1) PIWI.ip |
(9) TESTES |
| GTGCAGTCGAATGCCCCCTCTTTCCTTGTGGGCTGGATGGGTCCTTGATAGTATCCTTTTGTGCTCTTATCTATGGCTGTTCCTCTCCTGTTGGTTACTTTTCCTCTCTCAAACTGTTTCCTTCAAATCCTCATATCTGACATTGTAGTCTTTATTATTTAAGCTGTATAAATGAGAACGCACTTCATCAGCATGCTTCCTTAAAAGAAAAAAAAAACAGGTTCTGTCTTCTCGAAATTCATCATCTCCCTCATCTTTCTGAACACCTTT ............................................................((((((((((.(((((((..........((..((..((.................))..))..)).............(((......)))...........)))))))...))))))..))))....................................................................................... ..................................................51....................................................................................................................................185................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATCCTTTTGTGCTCTTATCTATGGCTGTTCCTCTCCTGTTGGTTACTTTT........................................................................................................................................................................ | 52 | 1 | 306.00 | 306.00 | 197.00 | 109.00 | - | - | - | - | - | - | - |
| .......................CCTTGTGGGCTGGATGGGTCCTTGAT............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TCTTCTCGAAATTCATCATCTCC..................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................................................................ATCATCTCCCTCATCTTTCTGAACA..... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................TATTTAAGCTGTATAAATGAGAACGC......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................AAGCTGTATAAATGAGAA............................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ......TCGAATGCCCCCTCTggag..................................................................................................................................................................................................................................................... | 19 | ggag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |
| GTGCAGTCGAATGCCCCCTCTTTCCTTGTGGGCTGGATGGGTCCTTGATAGTATCCTTTTGTGCTCTTATCTATGGCTGTTCCTCTCCTGTTGGTTACTTTTCCTCTCTCAAACTGTTTCCTTCAAATCCTCATATCTGACATTGTAGTCTTTATTATTTAAGCTGTATAAATGAGAACGCACTTCATCAGCATGCTTCCTTAAAAGAAAAAAAAAACAGGTTCTGTCTTCTCGAAATTCATCATCTCCCTCATCTTTCTGAACACCTTT ............................................................((((((((((.(((((((..........((..((..((.................))..))..)).............(((......)))...........)))))))...))))))..))))....................................................................................... ..................................................51....................................................................................................................................185................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................TCCTCTCCTGTTGGTTACTTTTCCTCTCTCA............................................................................................................................................................... | 31 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................TCTCCCTCATCTTTcatt............ | 18 | catt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - |