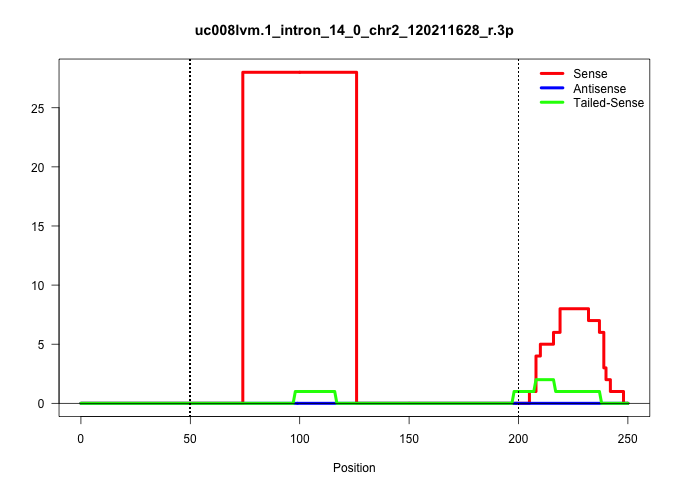
| Gene: Tmem87a | ID: uc008lvm.1_intron_14_0_chr2_120211628_r.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(11) TESTES |
| GGGACTATGTTGTATCAGTTCATGATCTTGCCTACGTTGCTGTGTGGTGTATAAGCTTGTCTTGCTTGTGATGACCATTTCTGTTACTGAAAGCTGAGGACTTTCTGGGCTGTATCCTGAAGCGATGTTCCATACTGCATTACTAACTTGTGCTCTAGATTCTTGAGACTTGAAGTTTTGATGCCCCTTTTTCTCCCTAGGCAATGCATGAACCATTGCAGACTTGGCAAGATGCTCCATACATTTTTAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................CCATTTCTGTTACTGAAAGCTGAGGACTTTCTGGGCTGTATCCTGAAGCGAT............................................................................................................................ | 52 | 1 | 28.00 | 28.00 | 28.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TGAACCATTGCAGACTTGGCAAGATGCTC............. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GCATGAACCATTGCAGACTTGGCAAGA.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................................................................................AACCATTGCAGACTTGGCAAGATGCTCCA........... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGACTTGGCAAGATGCTCCA........... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGCAGACTTGGCAAGATGCTCCATAC........ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................GACTTTCTGGGCTGTAgtg..................................................................................................................................... | 19 | gtg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGACTTGGCAAGATGCTCCATACATTTTT.. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TGAACCATTGCAGACTTGGCAAGATGCTCt............ | 30 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................................TGAACCATTGCAGACTTGGCAAGATGCTCCAT.......... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................................................................................................AGGCAATGCATGAACtgtg................................. | 19 | tgtg | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................................................................................TGAACCATTGCAGACTTGGCAAGATGCTCCA........... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| GGGACTATGTTGTATCAGTTCATGATCTTGCCTACGTTGCTGTGTGGTGTATAAGCTTGTCTTGCTTGTGATGACCATTTCTGTTACTGAAAGCTGAGGACTTTCTGGGCTGTATCCTGAAGCGATGTTCCATACTGCATTACTAACTTGTGCTCTAGATTCTTGAGACTTGAAGTTTTGATGCCCCTTTTTCTCCCTAGGCAATGCATGAACCATTGCAGACTTGGCAAGATGCTCCATACATTTTTAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|