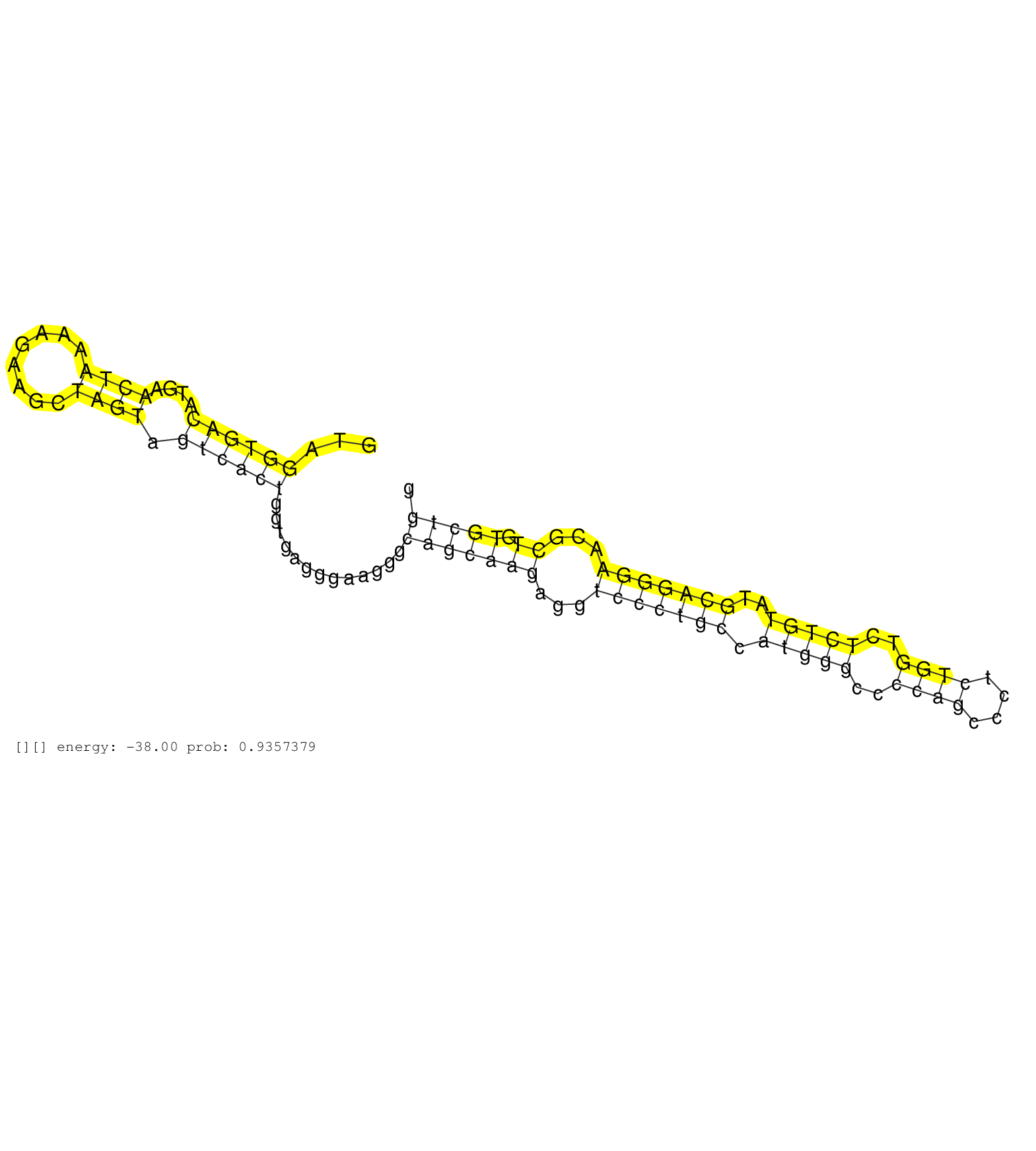

| Gene: Pla2g4b | ID: uc008luy.1_intron_4_0_chr2_119857139_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(7) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(23) TESTES |
| GAAACACTTCTTGTTACATCCACCCAGCGACCGGCCCTTCATCCCTTACAGTAGGTGACATGAACTAAAAGAAGCTAGTAGTCACTGGTGAGGGAAGGGCAGCAAGAGGTCCCTGCCATGGGCCCCAGCCCTCTGGTCTCTGTATGCAGGGAACGCTGTGCTGGTCATGGTGAGAGTGCCTGACAGGCTGTTGGCCCCTTGTCTCCCTGACCTTGCCTTCAGATCTCTACACACCAGCAACCTACCAGCTGACTGAAGAGGGCACTTTTAGG .....................................................((((((....((((........)))).)))))).............(((((((...(((((((.(((((..((((....))))..)))))..)))))))...)).)))))............................................................................................................. ..................................................51...............................................................................................................164.......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR037903(GSM510439) testes_rep4. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGTGACATGAACTAAAAGAAGCTAGT................................................................................................................................................................................................. | 29 | 1 | 14.00 | 14.00 | 2.00 | - | 4.00 | 2.00 | 1.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTAGGTGACATGAACTAAAAGAAGCTA................................................................................................................................................................................................... | 27 | 1 | 14.00 | 14.00 | 4.00 | 5.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................TACCAGCTGACTGAAGAGGGCACTTTT... | 27 | 1 | 12.00 | 12.00 | 2.00 | 2.00 | 2.00 | - | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTAGGTGACATGAACTAAAAGAAGCTAG.................................................................................................................................................................................................. | 28 | 1 | 11.00 | 11.00 | 3.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGACCTTGCCTTCAGATCTCTACACACC..................................... | 28 | 1 | 5.00 | 5.00 | - | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................TACCAGCTGACTGAAGAGGGCACTTT.... | 26 | 1 | 5.00 | 5.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGACATGAACTAAAAGAAGC..................................................................................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGACATGAACTAAAAGAAGCT.................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGTAGTCACTGGTGAGGGAAGGGCAGC......................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGACCTTGCCTTCAGATCTCTACACACCA.................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................TACCAGCTGACTGAAGAGGGCACTTTTA.. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGGTCTCTGTATGCAGGGAACGCTGTG................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGGTGAGAGTGCCTcatt....................................................................................... | 18 | catt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................CCAGCTGACTGAAGAGGGCACT...... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGACATGAACTAAAAGAAGCTAGTA................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................ACCTACCAGCTGACTGAAGAGGGCACTT..... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TTCATCCCTTACAGaagg......................................................................................................................................................................................................................... | 18 | aagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............TTACATCCACCCAGCGACCGGCCCTTCATCCCT.................................................................................................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGACATGAACTAAAAGAAGCTAGTAGTC............................................................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................CTGAAGAGGGCACTTTgt.. | 18 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................AACTAAAAGAAGCTAGTAGTCACTGGTG...................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................TGACCTTGCCTTCAGATCTCTACACACCAGC.................................. | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGACATGAACTAAAAGAAG...................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CCAGCGACCGGCCCTTCATCCCTTACA.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................AAAGAAGCTAGTAGTCACTGGTGAGGG.................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CCACCCAGCGACgcca............................................................................................................................................................................................................................................. | 16 | gcca | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGACCTTGCCTTCAGATCTCTACACACCAG................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................CCTACCAGCTGACTGAAGAGGGC......... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................AGCGACCGGCCCTTCATCCCTTACAGga........................................................................................................................................................................................................................... | 28 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGTGACATGAACTAAAAGAAGCTAGT................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGACATGAACTAAAAGAAGC..................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GAAACACTTCTTGTTACATCCACCCAGCGACCGGCCCTTCATCCCTTACAGTAGGTGACATGAACTAAAAGAAGCTAGTAGTCACTGGTGAGGGAAGGGCAGCAAGAGGTCCCTGCCATGGGCCCCAGCCCTCTGGTCTCTGTATGCAGGGAACGCTGTGCTGGTCATGGTGAGAGTGCCTGACAGGCTGTTGGCCCCTTGTCTCCCTGACCTTGCCTTCAGATCTCTACACACCAGCAACCTACCAGCTGACTGAAGAGGGCACTTTTAGG .....................................................((((((....((((........)))).)))))).............(((((((...(((((((.(((((..((((....))))..)))))..)))))))...)).)))))............................................................................................................. ..................................................51...............................................................................................................164.......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR037903(GSM510439) testes_rep4. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TCTGTATGCAGGGAaccc........................................................................................................................ | 18 | accc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |