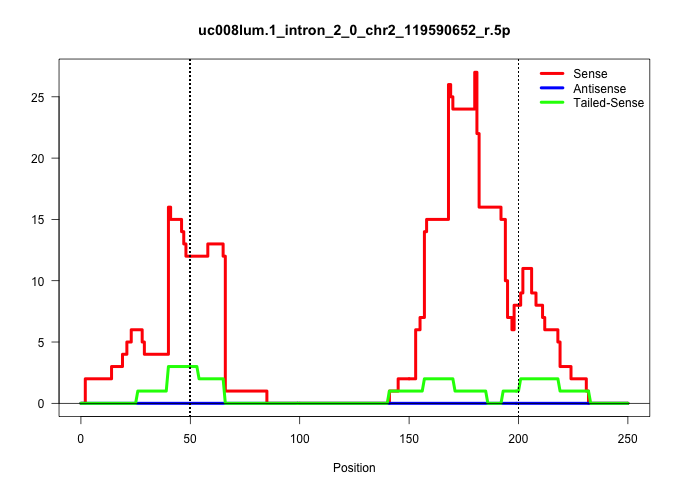
| Gene: mKIAA1403 | ID: uc008lum.1_intron_2_0_chr2_119590652_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(2) PIWI.mut |
(25) TESTES |
| TCTACCACCGAGCTTCAGATACTCCCTCTGGGCCACCTGCTGCTGACACTGTAGGCGTTGCCATGCGGGTCCTACAATGGGTGCTAGTTCTGGAGAGCTGGCGCCCCGAGGTCCTCTGGGCTGTGCCCCCTGCTGCCCGCCTAGCACGTCTCATGTGTGTGTACCTGGTGGATAGCGAGCTGTTTCGAGAGACTCCCATACAGCGACTGGTGGCAGCTCTTTTAGCTCGGCTCTGTCGGCCCCAAGTCTT .....................................................((((..(((....(((.(((.....((((((((((((....))))))))))))..))).)))...)))..))))...(((((.(((((..(((((.......)))))......))))).)))))......................................................................... .....................................................54..................................................................................................................................186.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................TGCTGACACTGTAGGCGTTGCCATGC........................................................................................................................................................................................ | 26 | 1 | 14.00 | 14.00 | 1.00 | 4.00 | 1.00 | 1.00 | 1.00 | 3.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGTGTACCTGGTGGATAGCGAGCTG.................................................................... | 25 | 1 | 6.00 | 6.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGGATAGCGAGCTGTTTCGAGAGACT........................................................ | 26 | 1 | 5.00 | 5.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TGTGTGTGTACCTGGTGGATAGCGAGCT..................................................................... | 28 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGGATAGCGAGCTGTTTCGAGAGACTC....................................................... | 27 | 1 | 3.00 | 3.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................ATGGGTGCTAGTTCTGG............................................................................................................................................................. | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TGACACTGTAGGCGTTGCCATGCGG...................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTTTCGAGAGACTCCCATACAGCGA............................................ | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GCGACTGGTGGCAGCTCTTTTAGCTCGGC................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGTGTACCTGGTGGATAGCGAGCTGTTTa................................................................ | 29 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................................................TGGATAGCGAGCTGTTTCGAGAGACTCCC..................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................TCTGGGCCACCTGCTGCTGACACTGTAa.................................................................................................................................................................................................... | 28 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CTCCCTCTGGGCCACCTGCTGCTGA............................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TCAGATACTCCCTCTGGGCCACCTGCT................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TAGCACGTCTCATGTGTGTGTACCTGGT................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................TGCCATGCGGGTCCTACAATGGGTGCT..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..TACCACCGAGCTTCAGATACTCCCTCT............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................AGCGACTGGTGGCAGCTCTTTTAGCTCGGCTt................. | 32 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................TGTGTGTACCTGGTGGATAGCGAGCT..................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GTGTACCTGGTGGATAGCGAGCTGT................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..TACCACCGAGCTTCAGATACTCCCTC.............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TCCCATACAGCGACTGGTGGCAGCTC............................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TTCGAGAGACTCCCATACAGCGACTGGTG...................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGTGTACCTGGTGGATAGCGAGCTGT................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGGATAGCGAGCTGTTTCGAGAGAC......................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GCGACTGGTGGCAGCTCTTTTAGCTCGGCT.................. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ACGTCTCATGTGTGTGTACCTGGTG................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TCCCATACAGCGACTGGTGGCAGttc............................... | 26 | ttc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CCCTCTGGGCCACCTGCTGCTGACA.......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTTTCGAGAGACTCCCATACAGCGACT.......................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................TACAGCGACTGGTGGCAGCTCTTTTA.......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGCTGACACTGTAGGCGTTGCCATG......................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TACTCCCTCTGGGCCACCTGCTGCTGAC........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................AGCGACTGGTGGCAGCT................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................TACAGCGACTGGTGGCAGCTC............................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGGATAGCGAGCTGTTTCGAGAGA.......................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TTCGAGAGACTCCCATACAGCGACTGGT....................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGCTGACACTGTAGGCGTTGCCctgc........................................................................................................................................................................................ | 26 | ctgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGCTGACACTGTAGGCGTTGCCgtgc........................................................................................................................................................................................ | 26 | gtgc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TAGCACGTCTCATGTGTGTGTACCTGGTGt............................................................................... | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| TCTACCACCGAGCTTCAGATACTCCCTCTGGGCCACCTGCTGCTGACACTGTAGGCGTTGCCATGCGGGTCCTACAATGGGTGCTAGTTCTGGAGAGCTGGCGCCCCGAGGTCCTCTGGGCTGTGCCCCCTGCTGCCCGCCTAGCACGTCTCATGTGTGTGTACCTGGTGGATAGCGAGCTGTTTCGAGAGACTCCCATACAGCGACTGGTGGCAGCTCTTTTAGCTCGGCTCTGTCGGCCCCAAGTCTT .....................................................((((..(((....(((.(((.....((((((((((((....))))))))))))..))).)))...)))..))))...(((((.(((((..(((((.......)))))......))))).)))))......................................................................... .....................................................54..................................................................................................................................186.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................TGGCGCCCCGAGGTCcgg......................................................................................................................................... | 18 | cgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TTTAGCTCGGCTCTtga................ | 17 | tga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |