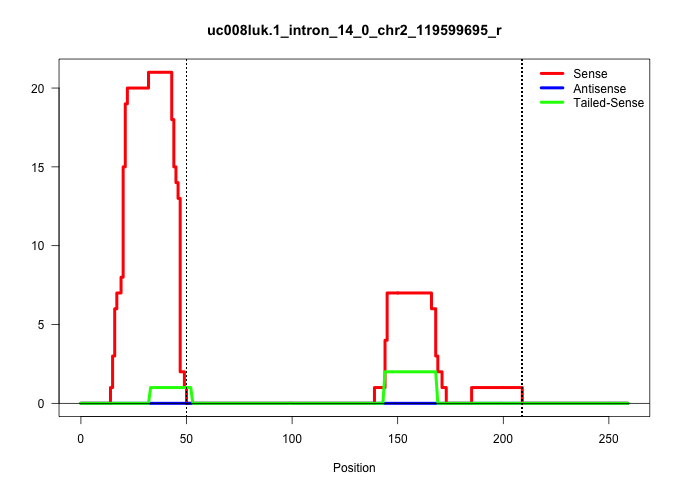
| Gene: Rpap1 | ID: uc008luk.1_intron_14_0_chr2_119599695_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(6) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(21) TESTES |
| CCCAGCAGAGAGCACTAGCATTGCAAGTGTTGTCCCAGATCGTCGGCAGGGTAAGTGGGCTGCTGGCCTGGTCCTACCAAGCCACACCCCATCCTTCATCCCTTGATCTTAGCCTTTTCCTCCTTACTGTGGGTCAGACTAGGAATGGGGCAAATGTGCAGTAACGGGTCAGCAAAGGTTCTATCCAAGTTCTGCATCTTGTACCCAAGGCCCAGGCTGGTGAGTTTGGGGACCGCCTAGTGGGCAGTGTCTTGCGCCT .................................................................................................................................................((((((((..((((((.(((((((.(((....)))..))))..))))))))).)))).)))).................................................... ...........................................................................................................................................140..................................................................209................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACGGG........................................................................................... | 24 | 1 | 10.00 | 10.00 | 7.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................CCTTACTGTGGGTCAGACTAGGAATGGGGCA........................................................................................................... | 31 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TTGCAAGTGTTGTCCCAGATCGTCGGC.................................................................................................................................................................................................................... | 27 | 1 | 6.00 | 6.00 | - | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................TGCAAGTGTTGTCCCAGATCGTCGGC.................................................................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACGGtt.......................................................................................... | 25 | tt | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACGGa........................................................................................... | 24 | a | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ................AGCATTGCAAGTGTTGTCCCAGATCGT........................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAGGAATGGGGCAAATGTGCAGTAACG............................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................CAAGTTCTGCATCTTGTACCCAAG.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................AAGTGTTGTCCCAGATCGTCG...................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CTAGCATTGCAAGTGTTGTCCCAGATCGTC....................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TTGCAAGTGTTGTCCCAGATCGTCGG..................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGGGCAAATGTGCAGTAACGGGTCA........................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TCCCAGATCGTCGGCAGG................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGGGCAAATGTGCAGTAACGGGT.......................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............TAGCATTGCAAGTGTTGTCCCAGATCGTCG...................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................GCAAGTGTTGTCCCAGATCGTCGGCAG.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................CCCAGATCGTCGGCAGGGTA.............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................AGCATTGCAAGTGTTGTCCCAGATCGTC....................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................ATTGCAAGTGTTGTCCCAGATCGTCGGC.................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGGGCAAATGTGCAGTAACGGGTCAGC...................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................CCCAGATCGTCGGCAGGGcc.............................................................................................................................................................................................................. | 20 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............TAGCATTGCAAGTGTTGTCCCAGATCGT........................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GCATTGCAAGTGTTGTCCCAGATCGTC....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| CCCAGCAGAGAGCACTAGCATTGCAAGTGTTGTCCCAGATCGTCGGCAGGGTAAGTGGGCTGCTGGCCTGGTCCTACCAAGCCACACCCCATCCTTCATCCCTTGATCTTAGCCTTTTCCTCCTTACTGTGGGTCAGACTAGGAATGGGGCAAATGTGCAGTAACGGGTCAGCAAAGGTTCTATCCAAGTTCTGCATCTTGTACCCAAGGCCCAGGCTGGTGAGTTTGGGGACCGCCTAGTGGGCAGTGTCTTGCGCCT .................................................................................................................................................((((((((..((((((.(((((((.(((....)))..))))..))))))))).)))).)))).................................................... ...........................................................................................................................................140..................................................................209................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................AAAGGTTCTATCCAtctg........................................................................ | 18 | tctg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................GTCCCAGATCGTCGtca...................................................................................................................................................................................................................... | 17 | tca | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................CAGATCGTCGGaata..................................................................................................................................................................................................................... | 15 | aata | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GTTGTCCCAGATCGTggat........................................................................................................................................................................................................................ | 19 | ggat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |